The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions
- PMID: 11483958
- PMCID: PMC3229202
- DOI: 10.1038/35087045
The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions
Abstract
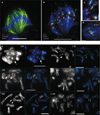
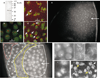
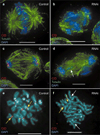
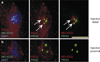
Centromere function requires the coordination of many processes including kinetochore assembly, sister chromatid cohesion, spindle attachment and chromosome movement. Here we show that CID, the Drosophila homologue of the CENP-A centromere-specific H3-like proteins, colocalizes with molecular-genetically defined functional centromeres in minichromosomes. Injection of CID antibodies into early embryos, as well as RNA interference in tissue-culture cells, showed that CID is required for several mitotic processes. Deconvolution fluorescence microscopy showed that CID chromatin is physically separate from proteins involved in sister cohesion (MEI-S332), centric condensation (PROD), kinetochore function (ROD, ZW10 and BUB1) and heterochromatin structure (HP1). CID localization is unaffected by mutations in mei-S332, Su(var)2-5 (HP1), prod or polo. Furthermore, the localization of POLO, CENP-meta, ROD, BUB1 and MEI-S332, but not PROD or HP1, depends on the presence of functional CID. We conclude that the centromere and flanking heterochromatin are physically and functionally separable protein domains that are required for different inheritance functions, and that CID is required for normal kinetochore formation and function, as well as cell-cycle progression.
Figures



References
-
- Dobie KW, Hari KL, Maggert KA, Karpen GH. Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev. 1999;9:206–217. - PubMed
-
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. - PubMed
-
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. - PubMed
-
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

