Identification and functional consequences of a new mutation (E155G) in the gene for GCAP1 that causes autosomal dominant cone dystrophy
- PMID: 11484154
- PMCID: PMC1235478
- DOI: 10.1086/323265
Identification and functional consequences of a new mutation (E155G) in the gene for GCAP1 that causes autosomal dominant cone dystrophy
Abstract
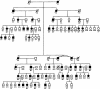
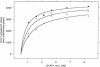
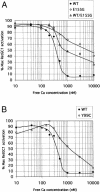
Mutations in the gene for guanylate cyclase-activating protein-1 (GCAP1) (GUCA1A) have been associated with autosomal dominant cone dystrophy (COD3). In the present study, a severe disease phenotype in a large white family was initially shown to map to chromosome 6p21.1, the location of GUCA1A. Subsequent single-stranded conformation polymorphism analysis and direct sequencing revealed an A464G transition, causing an E155G substitution within the EF4 domain of GCAP1. Modeling of the protein structure shows that the mutation eliminates a bidentate amino acid side chain essential for Ca2+ binding. This represents the first disease-associated mutation in GCAP1, or any neuron-specific calcium-binding protein within an EF-hand domain, that directly coordinates Ca2+. The functional consequences of this substitution were investigated in an in vitro assay of retinal guanylate cyclase activation. The mutant protein activates the cyclase at low Ca2+ concentrations but fails to inactivate at high Ca2+ concentrations. The overall effect of this would be the constitutive activation of guanylate cyclase in photoreceptors, even at the high Ca2+ concentrations of the dark-adapted state, which may explain the dominant disease phenotype.
Figures


References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for autosomal dominant retinal cone dystrophy, COD3, and GCAP1 [OMIM 180020, 602093, and 600364])
-
- Swiss-Model, http://www.expasy.ch/swissmod/SWISS-MODEL.html (for protein modeling)
References
-
- Balciuniene J, Johanssen K, Sandgren O, Wachmeister L, Homgren G, Forsman G (1995) A gene for autosomal dominant progressive cone dystrophy (CORD5) maps to chromosome 17p12-p13. Genomics 30:281–286 - PubMed
-
- Cuenca N, Lopez S, Howes K, Kolb H (1998) The localization of guanylyl cyclase-activating proteins in the mammalian retina. Invest Ophthalmol Vis Sci 39:1243–1250 - PubMed
-
- Dizhoor AM, Boikov SG, Olshevskaya EV (1998) Constitutive activation of photoreceptor guanylate cyclase by Y99C mutant of GCAP-1. J Biol Chem 273:17311–17314 - PubMed
-
- Dizhoor AM, Hurley JB (1999) Regulation of photoreceptor membrane guanylate cyclases by guanylate cyclase activator proteins. Methods 19:521–531 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

