Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas
- PMID: 11485924
- PMCID: PMC1850553
- DOI: 10.1016/S0002-9440(10)61737-3
Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas
Abstract
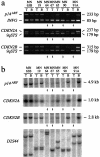
We investigated 67 meningothelial tumors (20 benign meningiomas, 34 atypical meningiomas, and 13 anaplastic meningiomas) for losses of genetic information from chromosome arms 1p and 9p, as well as for deletion, mutation, and expression of the tumor suppressor genes CDKN2A (p16(INKa)/MTS1), p14(ARF), CDKN2B (p15(INK4b)/MTS2) (all located at 9p21) and CDKN2C (1p32). Comparative genomic hybridization and microsatellite analysis showed losses on 1p in 11 anaplastic meningiomas (85%), 23 atypical meningiomas (68%), and 5 benign meningiomas (25%). One atypical meningioma with loss of heterozygosity on 1p carried a somatic CDKN2C mutation (c.202C>T: R68X). Losses on 9p were found in five anaplastic meningiomas (38%), six atypical meningiomas (18%), and one benign meningioma (5%). Six anaplastic meningiomas (46%) and one atypical meningioma (3%) showed homozygous deletions of the CDKN2A, p14(ARF), and CDKN2B genes. Two anaplastic meningiomas carried somatic point mutations in CDKN2A (c.262G>T: E88X and c.262G>A: E88K) and p14(ARF) (c.305G>T: G102V and c.305G>A: G102E). One anaplastic meningioma, three atypical meningiomas, and one benign meningioma without a demonstrated homozygous deletion or mutation of CDKN2A, p14(ARF), or CDKN2B lacked detectable transcripts from at least one of these genes. Hypermethylation of CDKN2A, p14(ARF), and CDKN2B could be demonstrated in one of these cases. Taken together, our results indicate that CDKN2C is rarely altered in meningiomas. However, the majority of anaplastic meningiomas either show homozygous deletions of CDKN2A, p14(ARF), and CDKN2B, mutations in CDKN2A and p14(ARF), or lack of expression of one or more of these genes. Thus, inactivation of the G(1)/S-phase cell-cycle checkpoint is an important aberration in anaplastic meningiomas.
Figures




References
-
- Louis DN, Scheithauer BW, Budka H, von Deimling A, Kepes JJ: Meningiomas. Pathology and Genetics of Tumours of the Nervous System. World Health Organization Classification of Tumours. Edited by P Kleihues, WK Cavenee. Lyon, IARC Press, 2000, pp 176–184
-
- Perry A, Scheithauer BW, Stafford SL, Lohse CM, Wollan PC: “Malignancy” in meningiomas: a clinicopathologic study of 116 patients, with grading implications. Cancer 1999, 85:2046-2056 - PubMed
-
- Ruttledge MH, Sarrazin J, Rangaratnam S, Phelan CM, Twist E, Merel P, Delattre O, Thomas G, Nordenskjöld M, Collins VP, Dumanski JP, Rouleau GA: Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet 1994, 6:180-184 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

