BIG: a calossin-like protein required for polar auxin transport in Arabidopsis
- PMID: 11485992
- PMCID: PMC312751
- DOI: 10.1101/gad.905201
BIG: a calossin-like protein required for polar auxin transport in Arabidopsis
Abstract
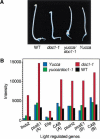
Polar auxin transport is crucial for the regulation of auxin action and required for some light-regulated responses during plant development. We have found that two mutants of Arabidopsis-doc1, which displays altered expression of light-regulated genes, and tir3, known for its reduced auxin transport-have similar defects and define mutations in a single gene that we have renamed BIG. BIG is very similar to the Drosophila gene Calossin/Pushover, a member of a gene family also present in Caenorhabditis elegans and human genomes. The protein encoded by BIG is extraordinary in size, 560 kD, and contains several putative Zn-finger domains. Expression-profiling experiments indicate that altered expression of multiple light-regulated genes in doc1 mutants can be suppressed by elevated levels of auxin caused by overexpression of an auxin biosynthetic gene, suggesting that normal auxin distribution is required to maintain low-level expression of these genes in the dark. Double mutants of tir3 with the auxin mutants pin1, pid, and axr1 display severe defects in auxin-dependent growth of the inflorescence. Chemical inhibitors of auxin transport change the intracellular localization of the auxin efflux carrier PIN1 in doc1/tir3 mutants, supporting the idea that BIG is required for normal auxin efflux.
Figures





References
-
- Anderson JT, Rogers RP, Jarrett HW. Ca2+-calmodulin binds to the carboxyl-terminal domain of dystrophin. J Biol Chem. 1996;271:6605–6610. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons; 1994.
-
- Bartel B, Fink GR. ILR1: An amidohydrolase that releases active indole-3-acetic acid from conjugates. Science. 1995;268:1745–1748. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
