Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex
- PMID: 11486016
- PMCID: PMC87296
- DOI: 10.1128/MCB.21.17.5767-5777.2001
Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex
Abstract
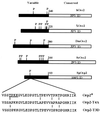
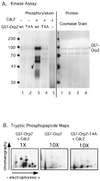
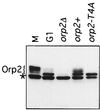
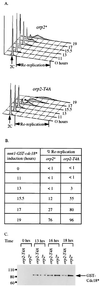
Cdc2 kinase is a master regulator of cell cycle progression in the fission yeast Schizosaccharomyces pombe. Our data indicate that Cdc2 phosphorylates replication factor Orp2, a subunit of the origin recognition complex (ORC). Cdc2 phosphorylation of Orp2 appears to be one of multiple mechanisms by which Cdc2 prevents DNA rereplication in a single cell cycle. Cdc2 phosphorylation of Orp2 is not required for Cdc2 to activate DNA replication initiation. Phosphorylation of Orp2 appears first in S phase and becomes maximal in G(2) and M when Cdc2 kinase activity is required to prevent reinitiation of DNA replication. A mutant lacking Cdc2 phosphorylation sites in Orp2 (orp2-T4A) allowed greater rereplication of DNA than congenic orp2 wild-type strains when the limiting replication initiation factor Cdc18 was deregulated. Thus, Cdc2 phosphorylation of Orp2 may be redundant with regulation of Cdc18 for preventing reinitiation of DNA synthesis. Since Cdc2 phosphorylation sites are present in Orp2 (also known as Orc2) from yeasts to metazoans, we propose that cell cycle-regulated phosphorylation of the ORC provides a safety net to prevent DNA rereplication and resulting genetic instability.
Figures

Similar articles
-
Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein.Nature. 1996 Jan 25;379(6563):360-3. doi: 10.1038/379360a0. Nature. 1996. PMID: 8552194
-
Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication.Cell. 2002 Nov 1;111(3):419-31. doi: 10.1016/s0092-8674(02)01042-5. Cell. 2002. PMID: 12419251
-
Negative regulation of Cdc18 DNA replication protein by Cdc2.Mol Biol Cell. 1998 Jan;9(1):63-73. doi: 10.1091/mbc.9.1.63. Mol Biol Cell. 1998. PMID: 9436991 Free PMC article.
-
The cdc18 protein initiates DNA replication in fission yeast.Prog Cell Cycle Res. 1997;3:135-42. doi: 10.1007/978-1-4615-5371-7_11. Prog Cell Cycle Res. 1997. PMID: 9552412 Review.
-
Cell cycle regulation in Schizosaccharomyces pombe.Curr Opin Microbiol. 2000 Dec;3(6):631-6. doi: 10.1016/s1369-5274(00)00152-1. Curr Opin Microbiol. 2000. PMID: 11121785 Review.
Cited by
-
Polo-like kinase 1 (Plk1): an Unexpected Player in DNA Replication.Cell Div. 2012 Feb 6;7:3. doi: 10.1186/1747-1028-7-3. Cell Div. 2012. PMID: 22309699 Free PMC article.
-
Site-specific interaction of the murine pre-replicative complex with origin DNA: assembly and disassembly during cell cycle transit and differentiation.Nucleic Acids Res. 2007;35(20):6701-13. doi: 10.1093/nar/gkm555. Epub 2007 Oct 4. Nucleic Acids Res. 2007. PMID: 17916579 Free PMC article.
-
A yeast GSK-3 kinase Mck1 promotes Cdc6 degradation to inhibit DNA re-replication.PLoS Genet. 2012;8(12):e1003099. doi: 10.1371/journal.pgen.1003099. Epub 2012 Dec 6. PLoS Genet. 2012. PMID: 23236290 Free PMC article.
-
Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant.Mol Biol Cell. 2006 May;17(5):2401-14. doi: 10.1091/mbc.e05-11-1043. Epub 2006 Feb 15. Mol Biol Cell. 2006. PMID: 16481397 Free PMC article.
-
Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts.Genes Dev. 2005 Jan 1;19(1):114-26. doi: 10.1101/gad.1255805. Epub 2004 Dec 14. Genes Dev. 2005. PMID: 15598982 Free PMC article.
References
-
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Berry L D, Gould K L. Regulation of Cdc2 activity by phosphorylation at T14/Y15. Prog Cell Cycle Res. 1996;2:99–105. - PubMed
-
- Booher R N, Alfa C E, Hyams J S, Beach D H. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
