Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat
- PMID: 11487690
- PMCID: PMC139131
- DOI: 10.1105/tpc.010083
Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat
Abstract
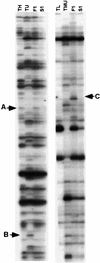
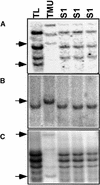
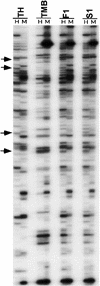
Interspecific or intergeneric hybridization, followed by chromosome doubling, can lead to the formation of new allopolyploid species. Recent studies indicate that allopolyploid formation is associated with genetic and epigenetic changes, although little is known about the type of changes that occur, how rapidly they occur, and the type of sequences involved. To address these matters, we have surveyed F1 hybrids between diploid species from the wheat (Aegilops and Triticum) group and their derived allotetraploids by screening a large number of loci using amplified fragment length polymorphism and DNA gel blot analysis and by assaying the extent of cytosine methylation. We found that sequence elimination is one of the major and immediate responses of the wheat genome to wide hybridization or allopolyploidy, that it affects a large fraction of the genome, and that it is reproducible. In one cross between AE: sharonensis x AE: umbellulata, 14% of the loci from AE: sharonensis were eliminated compared with only 0.5% from AE: umbellulata, with most changes occurring in the F1 hybrid. In contrast, crosses between AE: longissima x T. urartu showed that sequence elimination was more frequent after chromosome doubling. Alterations in cytosine methylation occurred in approximately 13% of the loci, either in the F1 hybrid or in the allopolyploid. For eight of nine bands that were isolated, the sequences that underwent elimination corresponded to low-copy DNA, whereas alterations in methylation patterns affected both repetitive DNA sequences, such as retrotransposons, and low-copy DNA in approximately equal proportions.
Figures
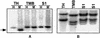
References
-
- The Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Banks, J.A., Masson, P., and Fedoroff, N. (1988). Molecular mechanisms in the developmental regulation of the maize Suppressor-mutator transposable element. Genes Dev. 2, 1364–1380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

