Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium
- PMID: 11487704
- PMCID: PMC139132
- DOI: 10.1105/tpc.010084
Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium
Abstract
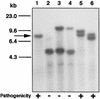
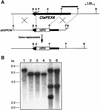
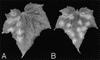
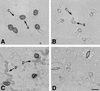
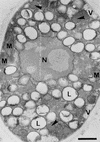
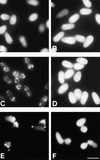
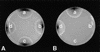
Peroxisomes are organelles that perform a wide range of metabolic functions in eukaryotic cells. However, their role in fungal pathogenesis is poorly understood. Here we report that ClaPEX6, an ortholog of PEX6, is required for the fungus Colletotrichum lagenarium to infect host plants. ClaPEX6 was identified in random insertional mutagenesis experiments aimed at elucidating genes involved in pathogenesis. Functional analysis, using a green fluorescent protein cassette containing the peroxisomal targeting signal1 (PTS1), revealed that import of PTS1-containing proteins is impaired in clapex6 mutants generated by targeted gene disruption. Failure of growth on fatty acids shows an inability of fatty acid beta-oxidation in these mutants. These results indicate that disruption of ClaPEX6 impairs peroxisomal metabolism, even though clapex6 mutants show normal growth and conidiation in nutrient-rich conditions. The clapex6 mutants formed small appressoria with severely reduced melanization that failed to form infectious hyphae. These data indicate that peroxisomes are necessary for appressorium-mediated penetration of host plants. The addition of glucose increased the pathogenicity of clapex6 mutants, suggesting that the glucose metabolic pathway can compensate partially for peroxisomes in phytopathogenicity.
Figures

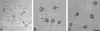
Similar articles
-
Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium.Appl Environ Microbiol. 2006 Sep;72(9):6345-54. doi: 10.1128/AEM.00988-06. Appl Environ Microbiol. 2006. PMID: 16957261 Free PMC article.
-
Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare.Mol Plant Microbe Interact. 2010 Apr;23(4):436-45. doi: 10.1094/MPMI-23-4-0436. Mol Plant Microbe Interact. 2010. PMID: 20192831
-
Colletotrichum orbiculare FAM1 Encodes a Novel Woronin Body-Associated Pex22 Peroxin Required for Appressorium-Mediated Plant Infection.mBio. 2015 Sep 15;6(5):e01305-15. doi: 10.1128/mBio.01305-15. mBio. 2015. PMID: 26374121 Free PMC article.
-
Function of peroxisomes in plant-pathogen interactions.Subcell Biochem. 2013;69:329-45. doi: 10.1007/978-94-007-6889-5_18. Subcell Biochem. 2013. PMID: 23821157 Review.
-
A newly discovered function of peroxisomes: involvement in biotin biosynthesis.Plant Signal Behav. 2012 Dec;7(12):1589-93. doi: 10.4161/psb.22405. Epub 2012 Oct 16. Plant Signal Behav. 2012. PMID: 23073000 Free PMC article. Review.
Cited by
-
Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein.Plant Cell. 2002 Aug;14(8):1869-83. doi: 10.1105/tpc.002576. Plant Cell. 2002. PMID: 12172028 Free PMC article.
-
From signal transduction to autophagy of plant cell organelles: lessons from yeast and mammals and plant-specific features.Protoplasma. 2010 Dec;247(3-4):233-56. doi: 10.1007/s00709-010-0190-0. Epub 2010 Aug 24. Protoplasma. 2010. PMID: 20734094 Review.
-
Unveiling the Role Displayed by Penicillium digitatum PdMut3 Transcription Factor in Pathogen-Fruit Interaction.J Fungi (Basel). 2021 Oct 3;7(10):828. doi: 10.3390/jof7100828. J Fungi (Basel). 2021. PMID: 34682249 Free PMC article.
-
Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis.Genome Biol. 2009;10(1):R4. doi: 10.1186/gb-2009-10-1-r4. Epub 2009 Jan 9. Genome Biol. 2009. PMID: 19134172 Free PMC article.
-
Mechanism Analysis of Amphotericin B Controlling Postharvest Gray Mold in Table Grapes.Foods. 2025 Apr 3;14(7):1260. doi: 10.3390/foods14071260. Foods. 2025. PMID: 40238543 Free PMC article.
References
-
- Agrios, G.N. (1988). Plant Pathology, 3rd ed. (San Diego, CA: Academic Press).
-
- Baerends, R.J.S., Faber, K.N., Kiel, J.A.K.W., van der Klei, J., Harder, W., and Veenhuis, M. (2000). Sorting and function of peroxisomal membrane proteins. FEMS Microbiol. Rev. 24, 291–301. - PubMed
-
- Bechinger, C., Giebel, K.-F., Schnell, M., Leiderer, P., Deising, H.B., and Bastmeyer, M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285, 1896–1899. - PubMed
-
- Beevers, H. (1979). Microbodies in higher plants. Annu. Rev. Plant Physiol. 30, 159–193.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

