Somatic hypermutation shapes the antibody repertoire of memory B cells in humans
- PMID: 11489956
- PMCID: PMC2193472
- DOI: 10.1084/jem.194.3.375
Somatic hypermutation shapes the antibody repertoire of memory B cells in humans
Abstract
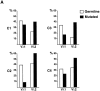
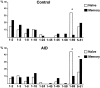
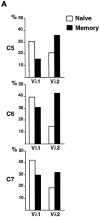
High-affinity antibodies produced by memory B cells differ from antibodies produced in naive B cells in two respects. First, many of these antibodies show somatic hypermutation, and second, the repertoire of antibodies expressed in memory responses is highly selected. To determine whether somatic hypermutation is responsible for the shift in the antibody repertoire during affinity maturation, we analyzed the immunoglobulin lambda light chain (Iglambda) repertoire expressed by naive and antigen-selected memory B cells in humans. We found that the Iglambda repertoire differs between naive and memory B cells and that this shift in the repertoire does not occur in the absence of somatic hypermutation in patients lacking activation-induced cytidine deaminase (AID). Our work suggests that somatic hypermutation makes a significant contribution to shaping the antigen-selected antibody repertoire in humans.
Figures




References
-
- Siskind G.W., Eisen H.N. Effect of variation in antibody-hapten association constant upon the biologic activity of the antibody. J. Immunol. 1965;95:436–441. - PubMed
-
- Weigert M.G., Cesari I.M., Yonkovich S.J., Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. - PubMed
-
- Griffiths G.M., Berek C., Kaartinen M., Milstein C. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature. 1984;312:271–275. - PubMed
-
- Kaartinen M., Griffiths G.M., Hamlyn P.H., Markham A.F., Karjalainen K., Pelkonen J.L., Makela O., Milstein C. Anti-oxazolone hybridomas and the structure of the oxazolone idiotype. J. Immunol. 1983;130:937–945. - PubMed
-
- Kaartinen M., Griffiths G.M., Markham A.F., Milstein C. mRNA sequences define an unusually restricted IgG response to 2-phenyloxazolone and its early diversification. Nature. 1983;304:320–324. - PubMed

