A metallothionein containing a zinc finger within a four-metal cluster protects a bacterium from zinc toxicity
- PMID: 11493688
- PMCID: PMC55497
- DOI: 10.1073/pnas.171120098
A metallothionein containing a zinc finger within a four-metal cluster protects a bacterium from zinc toxicity
Abstract
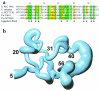
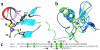
Zinc is essential for many cellular processes, including DNA synthesis, transcription, and translation, but excess can be toxic. A zinc-induced gene, smtA, is required for normal zinc-tolerance in the cyanobacterium Synechococcus PCC 7942. Here we report that the protein SmtA contains a cleft lined with Cys-sulfur and His-imidazole ligands that binds four zinc ions in a Zn(4)Cys(9)His(2) cluster. The thiolate sulfurs of five Cys ligands provide bridges between the two ZnCys(4) and two ZnCys(3)His sites, giving two fused six-membered rings with distorted boat conformations. The inorganic core strongly resembles the Zn(4)Cys(11) cluster of mammalian metallothionein, despite different amino acid sequences, a different linear order of the ligands, and presence of histidine ligands. Also, SmtA contains elements of secondary structure not found in metallothioneins. One of the two Cys(4)-coordinated zinc ions in SmtA readily exchanges with exogenous metal ((111)Cd), whereas the other is inert. The thiolate sulfur ligands bound to zinc in this site are buried within the protein. Regions of beta-strand and alpha-helix surround the inert site to form a zinc finger resembling the zinc fingers in GATA and LIM-domain proteins. Eukaryotic zinc fingers interact specifically with other proteins or DNA and an analogous interaction can therefore be anticipated for prokaryotic zinc fingers. SmtA now provides structural proof for the existence of zinc fingers in prokaryotes, and sequences related to the zinc finger motif can be identified in several bacterial genomes.
Figures

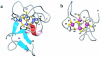
References
-
- Robinson N J, Gupta A, Fordham-Skelton A P, Croy R R D, Whitton B A, Huckle J W. Proc R Soc London Ser B. 1990;242:241–247. - PubMed
-
- Huckle J W, Morby A P, Turner J S, Robinson N J. Mol Microbiol. 1993;7:177–187. - PubMed
-
- Turner J S, Morby A P, Whitton A B, Gupta A, Robinson N J. J Biol Chem. 1993;268:4494–4498. - PubMed
-
- Bird A J, Turner-Cavet J S, Lakey J S, Robinson N J. J Biol Chem. 1998;273:21246–21252. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

