Hepatitis C virus replication is directly inhibited by IFN-alpha in a full-length binary expression system
- PMID: 11493707
- PMCID: PMC55541
- DOI: 10.1073/pnas.171319698
Hepatitis C virus replication is directly inhibited by IFN-alpha in a full-length binary expression system
Abstract
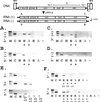
Hepatitis C virus (HCV) is a leading cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma. The absence of culture systems permissive for HCV replication has presented a major bottleneck to antiviral development. We sought to recapitulate the early steps in the life cycle of HCV by means of DNA-based expression of viral genomic sequences. Here we report expression of replicating HCV RNA by using a, to our knowledge, novel binary expression system in which cells were transfected with a T7 polymerase-driven full-length HCV cDNA plasmid containing a cis-acting hepatitis Delta ribozyme to control 3' cleavage, and infected with vaccinia-T7 polymerase. HCV genomic and replicative strand synthesis, in addition to protein synthesis, was detectable and depended on full-length HCV sequences. Moreover, the system was capable of generating HCV RNA quasispecies, consistent with the action of the low-fidelity HCV NS5B RNA polymerase. IFN-alpha, but not ribavirin, directly inhibited the viral replicative cycle in these cells, identifying the virus itself and not solely the immune system as a direct target of IFN action. The availability of a cell-based test for viral replication will facilitate screening of inhibitory compounds, analysis of IFN-resistance mechanisms, and analysis of virus-host cell interactions.
Figures


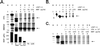
Similar articles
-
Hepatitis C virus replication is inhibited by 22beta-methoxyolean-12-ene-3beta, 24(4beta)-diol (ME3738) through enhancing interferon-beta.Hepatology. 2008 Jul;48(1):59-69. doi: 10.1002/hep.22289. Hepatology. 2008. PMID: 18459156
-
Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system.J Virol. 2002 Sep;76(17):8505-17. doi: 10.1128/jvi.76.17.8505-8517.2002. J Virol. 2002. PMID: 12163570 Free PMC article.
-
Interferon alpha-2b inhibits negative-strand RNA and protein expression from full-length HCV1a infectious clone.Exp Mol Pathol. 2004 Jun;76(3):242-52. doi: 10.1016/j.yexmp.2004.01.004. Exp Mol Pathol. 2004. PMID: 15126107
-
Molecular virology of hepatitis C virus: an update with respect to potential antiviral targets.Antivir Ther. 1998;3(Suppl 3):71-81. Antivir Ther. 1998. PMID: 10726057 Review.
-
Innate and adaptive immune responses in HCV infections.J Hepatol. 2014 Nov;61(1 Suppl):S14-25. doi: 10.1016/j.jhep.2014.06.035. Epub 2014 Nov 3. J Hepatol. 2014. PMID: 25443342 Review.
Cited by
-
Nanomedicines in the treatment of patients with hepatitis C co-infected with HIV--focus on pegylated interferon-alpha.Int J Nanomedicine. 2006;1(4):399-409. doi: 10.2147/nano.2006.1.4.399. Int J Nanomedicine. 2006. PMID: 17722274 Free PMC article. Review.
-
Alpha interferon inhibits hepatitis C virus replication in primary human hepatocytes infected in vitro.J Virol. 2002 Aug;76(16):8189-99. doi: 10.1128/jvi.76.16.8189-8199.2002. J Virol. 2002. PMID: 12134024 Free PMC article.
-
Genetic evolution of structural region of hepatitis C virus in primary infection.World J Gastroenterol. 2002 Aug;8(4):686-93. doi: 10.3748/wjg.v8.i4.686. World J Gastroenterol. 2002. PMID: 12174379 Free PMC article.
-
Subcellular forms and biochemical events triggered in human cells by HCV polyprotein expression from a viral vector.Virol J. 2008 Sep 15;5:102. doi: 10.1186/1743-422X-5-102. Virol J. 2008. PMID: 18793431 Free PMC article.
-
Human systems immunology: hypothesis-based modeling and unbiased data-driven approaches.Semin Immunol. 2013 Oct 31;25(3):193-200. doi: 10.1016/j.smim.2012.11.003. Epub 2013 Jan 29. Semin Immunol. 2013. PMID: 23375135 Free PMC article. Review.
References
-
- Alter M J, Kruszon-Moran D, Nainan O V, McQuillan G M, Gao F, Moyer L A, Kaslow R A, Margolis H S. N Engl J Med. 1999;341:556–562. - PubMed
-
- El-Serag H B, Mason A C. N Engl J Med. 1999;340:745–750. - PubMed
-
- Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N, et al. Science. 1992;258:135–140. - PubMed
-
- Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder J C, Strazzera A, Chien D Y, Munoz S J, Balestrieri A, et al. Science. 2000;288:339–344. - PubMed
-
- McHutchison J G, Poynard T. Semin Liver Dis. 1999;19:57–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

