The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB
- PMID: 11500365
- PMCID: PMC125563
- DOI: 10.1093/emboj/20.16.4380
The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB
Abstract
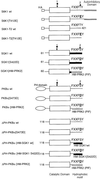
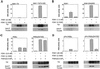
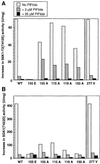
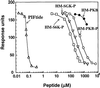
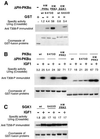
PKB/Akt, S6K1 and SGK are related protein kinases activated in a PI 3-kinase-dependent manner in response to insulin/growth factors signalling. Activation entails phosphorylation of these kinases at two residues, the T-loop and the hydrophobic motif. PDK1 activates S6K, SGK and PKB isoforms by phosphorylating these kinases at their T-loop. We demonstrate that a pocket in the kinase domain of PDK1, termed the 'PIF-binding pocket', plays a key role in mediating the interaction and phosphorylation of S6K1 and SGK1 at their T-loop motif by PDK1. Our data indicate that prior phosphorylation of S6K1 and SGK1 at their hydrophobic motif promotes their interaction with the PIF-binding pocket of PDK1 and their T-loop phosphorylation. Thus, the hydrophobic motif phosphorylation of S6K and SGK converts them into substrates that can be activated by PDK1. In contrast, the PIF-binding pocket of PDK1 is not required for the phosphorylation of PKBalpha by PDK1. The PIF-binding pocket represents a substrate recognition site on a protein kinase that is only required for the phosphorylation of a subset of its physiological substrates.
Figures


References
-
- Alessi D.R., Cohen,P., Ashworth,A., Cowley,S., Leevers,S.J. and Marshall,C.J. (1995) Assay and expression of mitogen-activated protein kinase, MAP kinase kinase and Raf. Methods Enzymol., 255, 279–290. - PubMed
-
- Alessi D.R., Deak,M., Casamayor,A. et al. (1997a) 3-phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol., 7, 776–789. - PubMed
-
- Alessi D.R., James,S.R., Downes,C.P., Holmes,A.B., Gaffney,P.R., Reese,C.B. and Cohen,P. (1997b) Characterization of a 3-phospho inositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol., 7, 261–269. - PubMed
-
- Alessi D.R., Kozlowski,M.T., Weng,Q.P., Morrice,N. and Avruch,J. (1998) 3-phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol., 8, 69–81. - PubMed
-
- Andjelkovic M., Alessi,D.R., Meier,R. et al. (1997) Role of translocation in the activation and function of protein kinase B. J. Biol. Chem., 272, 31515–31524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

