A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site
- PMID: 11500380
- PMCID: PMC125580
- DOI: 10.1093/emboj/20.16.4536
A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site
Abstract
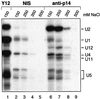
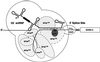
Previous UV cross-linking studies demonstrated that, upon integration of the U2 snRNP into the spliceosome, a 14 kDa protein (p14) interacts directly with the branch adenosine, the nucleophile for the first transesterification step of splicing. We have identified the cDNA encoding this protein by microsequencing a 14 kDa protein isolated from U2-type spliceosomes. This protein contains an RNA recognition motif and is highly conserved across species. Antibodies raised against this cDNA-encoded protein precipitated the 14 kDa protein cross-linked to the branch adenosine, confirming the identity of the p14 cDNA. A combination of immunoblotting, protein microsequencing and immunoprecipitation revealed that p14 is a component of both 17S U2 and 18S U11/U12 snRNPs, suggesting that it contributes to the interaction of these snRNPs with the branch sites of U2- and U12-type pre-mRNAs, respectively. p14 was also shown to be a subunit of the heteromeric splicing factor SF3b and to interact directly with SF3b155. Immuno precipitations indicated that p14 is present in U12-type spliceosomes, consistent with the idea that branch point selection is similar in the major and minor spliceosomes.
Figures





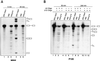
References
-
- Abovich N. and Rosbash,M. (1997) Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell, 89, 403–412. - PubMed
-
- Bennett M., Michaud,S., Kingston,J. and Reed,R. (1992) Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev., 6, 1986–2000. - PubMed
-
- Berglund J.A., Chua,K., Abovich,N., Reed,R. and Rosbash,M. (1997) The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell, 89, 781–787. - PubMed
-
- Brosi R., Hauri,H.P. and Kramer,A. (1993) Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem., 268, 17640–17646. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

