Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus
- PMID: 11500385
- PMCID: PMC125560
- DOI: 10.1093/emboj/20.16.4588
Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus
Abstract
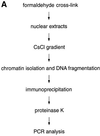
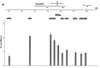
Epstein-Barr virus (EBV) replicates in its latent phase once per cell cycle in proliferating B cells. The latent origin of DNA replication, oriP, supports replication and stable maintenance of the EBV genome. OriP comprises two essential elements: the dyad symmetry (DS) and the family of repeats (FR), both containing clusters of binding sites for the transactivator EBNA1. The DS element appears to be the functional replicator. It is not yet understood how oriP-dependent replication is integrated into the cell cycle and how EBNA1 acts at the molecular level. Using chromatin immunoprecipitation experiments, we show that the human origin recognition complex (hsORC) binds at or near the DS element. The association of hsORC with oriP depends on the DS element. Deletion of this element not only abolishes hsORC binding but also reduces replication initiation at oriP to background level. Co-immunoprecipitation experiments indicate that EBNA1 is associated with hsORC in vivo. These results indicate that oriP might use the same cellular initiation factors that regulate chromosomal replication, and that EBNA1 may be involved in recruiting hsORC to oriP.
Figures








References
-
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. - PubMed
-
- Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

