CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells
- PMID: 11500386
- PMCID: PMC125570
- DOI: 10.1093/emboj/20.16.4603
CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells
Abstract
CENP-H has recently been discovered as a constitutive component of the centromere that co-localizes with CENP-A and CENP-C throughout the cell cycle. The precise function, however, remains poorly understood. We examined the role of CENP-H in centromere function and assembly by generating a conditional loss-of-function mutant in the chicken DT40 cell line. In the absence of CENP-H, cell cycle arrest at metaphase, consistent with loss of centromere function, was observed. Immunocytochemical analysis of the CENP-H-deficient cells demonstrated that CENP-H is necessary for CENP-C, but not CENP-A, localization to the centromere. These findings indicate that centromere assembly in vertebrate cells proceeds in a hierarchical manner in which localization of the centromere-specific histone CENP-A is an early event that occurs independently of CENP-C and CENP-H.
Figures







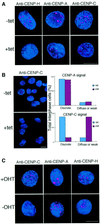
References
-
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. - PubMed
-
- Brown M.T. (1995) Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene, 160, 111–116. - PubMed
-
- Buerstedde J.M. and Takeda,S. (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell, 67, 179–188. - PubMed
-
- Chan G.K., Jablonski,S.A., Starr,D.A., Goldberg,M.L. and Yen,T.J. (2000) Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nature Cell Biol., 2, 944–947. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

