Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1
- PMID: 11500548
- PMCID: PMC117149
- DOI: 10.1104/pp.126.4.1493
Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1
Abstract
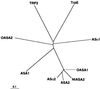
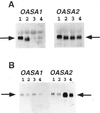
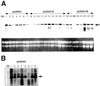
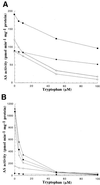
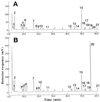
Anthranilate synthase (AS) is a key enzyme in the synthesis of tryptophan (Trp), indole-3-acetic acid, and indole alkaloids. Two genes, OASA1 and OASA2, encoding AS alpha-subunits were isolated from a monocotyledonous plant, rice (Oryza sativa cv Nipponbare), and were characterized. A phylogenetic tree of AS alpha-subunits from various species revealed a close evolutionary relationship among OASA1 and Arabidopsis ASA2, Ruta graveolens AS alpha 2, and tobacco ASA2, whereas OASA2, Arabidopsis ASA1, and R. graveolens AS alpha 1 were more distantly related. OASA1 is expressed in all tissues tested, but the amount of its mRNA was greater in panicles than in leaves and roots. The abundance of OASA2 transcripts is similar among tissues and greater than that of OASA1 transcripts; furthermore, OASA2 expression was induced by a chitin heptamer, a potent elicitor, suggesting that OASA2 participates in secondary metabolism. Expression of wild-type OASA1 or OASA2 transgenes did not affect the Trp content of rice calli or plants. However, transformed calli and plants expressing a mutated OASA1 gene, OASA1(D323N), that encodes a protein in which aspartate-323 is replaced with asparagine manifested up to 180- and 35-fold increases, respectively, in Trp accumulation. These transgenic calli and plants were resistant to 300 microM 5-methyl-Trp, and AS activity of the calli showed a markedly reduced sensitivity to Trp. These results show that OASA1 is important in the regulation of free Trp concentration, and that mutation of OASA1 to render the encoded protein insensitive to feedback inhibition results in accumulation of Trp at high levels. The OASA1(D323N) transgene may prove useful for the generation of crops with an increased Trp content.
Figures



References
-
- Bohlmann J, DeLuca V, Eilert U, Martin W. Purification and cDNA cloning of anthranilate synthase from Ruta graveolens: modes of expression and properties of native and recombinant enzymes. Plant J. 1995;7:491–501. - PubMed
-
- Caligiuri MG, Bauerle R. Identification of amino acid residues involved in feedback regulation of the anthranilate synthase complex from Salmonella typhimurium. J Biol Chem. 1991;266:8328–8335. - PubMed
-
- Chu C, Wang C, Sun C, Hsu C, Yin K, Chu C, Bi F. Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Scientia Sinica. 1975;18:223–231. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

