Phylogenetic relationships within cation transporter families of Arabidopsis
- PMID: 11500563
- PMCID: PMC117164
- DOI: 10.1104/pp.126.4.1646
Phylogenetic relationships within cation transporter families of Arabidopsis
Abstract
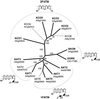
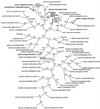
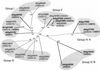
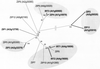
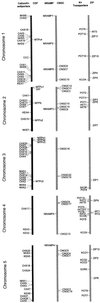
Uptake and translocation of cationic nutrients play essential roles in physiological processes including plant growth, nutrition, signal transduction, and development. Approximately 5% of the Arabidopsis genome appears to encode membrane transport proteins. These proteins are classified in 46 unique families containing approximately 880 members. In addition, several hundred putative transporters have not yet been assigned to families. In this paper, we have analyzed the phylogenetic relationships of over 150 cation transport proteins. This analysis has focused on cation transporter gene families for which initial characterizations have been achieved for individual members, including potassium transporters and channels, sodium transporters, calcium antiporters, cyclic nucleotide-gated channels, cation diffusion facilitator proteins, natural resistance-associated macrophage proteins (NRAMP), and Zn-regulated transporter Fe-regulated transporter-like proteins. Phylogenetic trees of each family define the evolutionary relationships of the members to each other. These families contain numerous members, indicating diverse functions in vivo. Closely related isoforms and separate subfamilies exist within many of these gene families, indicating possible redundancies and specialized functions. To facilitate their further study, the PlantsT database (http://plantst.sdsc.edu) has been created that includes alignments of the analyzed cation transporters and their chromosomal locations.
Figures




References
-
- Ache P, Becker D, Ivashikina N, Dietrich P, Roelfsema MR, Hedrich R. GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K(+)-selective, K(+)-sensing ion channel. FEBS Lett. 2000;486:93–98. - PubMed
-
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. - PubMed
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

