A novel superoxide dismutase with a high isoelectric point in higher plants. expression, regulation, and protein localization
- PMID: 11500564
- PMCID: PMC117165
- DOI: 10.1104/pp.126.4.1668
A novel superoxide dismutase with a high isoelectric point in higher plants. expression, regulation, and protein localization
Abstract
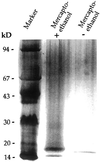
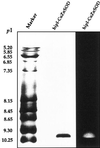
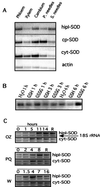
Several isoforms of superoxide dismutase (SOD) with a high isoelectric point (pI) have been identified by isoelectric focusing chromatography in protein extracts from Scots pine (Pinus sylvestris) needles. One of these isoforms, a CuZn-SOD with a pI of about 10 and thus denoted hipI-SOD, has been isolated and purified to apparent homogeneity. A cDNA encoding the hipI-SOD protein was cloned and sequenced. Northern hybridization of mRNA isolated from different organs and tissues showed that hipI-SOD has a markedly different pattern of expression compared with chloroplastic and cytosolic SOD. Furthermore, the transcript levels of hipI-SOD and cytosolic SOD were found to respond differently to mechanical wounding, treatment with oxidized glutathione, paraquat, and ozone. Immunogold electron microscopy localized the hipI-SOD in the plasma membrane of sieve cells and the Golgi apparatus of albuminous cells. Moreover, high protein density was also detected in extracellular spaces such as secondary cell wall thickenings of the xylem and sclerenchyma and in intercellular spaces of parenchyma cells.
Figures



Similar articles
-
A small family of novel CuZn-superoxide dismutases with high isoelectric points in hybrid aspen.Planta. 2001 Jun;213(2):272-9. doi: 10.1007/s004250000505. Planta. 2001. PMID: 11469593
-
Pinus sylvestris L. needles contain extracellular CuZn superoxide dismutase.Planta. 1994;192(2):195-201. Planta. 1994. PMID: 7764316
-
Characterization of cDNAs encoding CuZn-superoxide dismutases in Scots pine.Plant Mol Biol. 1992 Feb;18(3):545-55. doi: 10.1007/BF00040670. Plant Mol Biol. 1992. PMID: 1371406
-
Mouse extracellular superoxide dismutase: primary structure, tissue-specific gene expression, chromosomal localization, and lung in situ hybridization.Am J Respir Cell Mol Biol. 1997 Oct;17(4):393-403. doi: 10.1165/ajrcmb.17.4.2826. Am J Respir Cell Mol Biol. 1997. PMID: 9376114 Review.
-
The regulation and function of tobacco superoxide dismutases.Free Radic Biol Med. 1997;23(3):515-20. doi: 10.1016/s0891-5849(97)00112-3. Free Radic Biol Med. 1997. PMID: 9214590 Review.
Cited by
-
Superoxide dismutase, peroxidase, and germin-like protein activity in plasma membranes and apoplast of maize roots.Protoplasma. 2005 Dec;226(3-4):191-7. doi: 10.1007/s00709-005-0112-8. Epub 2005 Oct 26. Protoplasma. 2005. PMID: 16244808
-
Salicylic acid stimulates secretion of the normally symplastic enzyme mannitol dehydrogenase: a possible defense against mannitol-secreting fungal pathogens.Planta. 2009 Nov;230(6):1093-103. doi: 10.1007/s00425-009-1006-3. Epub 2009 Sep 1. Planta. 2009. PMID: 19727802
-
Ectopic lignification in the flax lignified bast fiber1 mutant stem is associated with tissue-specific modifications in gene expression and cell wall composition.Plant Cell. 2014 Nov;26(11):4462-82. doi: 10.1105/tpc.114.130443. Epub 2014 Nov 7. Plant Cell. 2014. PMID: 25381351 Free PMC article.
-
Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress.Plant Mol Biol. 2015 Apr;87(6):615-31. doi: 10.1007/s11103-015-0301-6. Epub 2015 Mar 10. Plant Mol Biol. 2015. PMID: 25754733
-
Production of reactive oxygen species by plant NADPH oxidases.Plant Physiol. 2006 Jun;141(2):336-40. doi: 10.1104/pp.106.078089. Plant Physiol. 2006. PMID: 16760484 Free PMC article. No abstract available.
References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols of Molecular Biology. New York: Green Publishing Associates/Wiley Interscience; 1989. pp. 6.0.3–6.4.7.
-
- Beauchamp CO, Fridovich I. Superoxide dismutase: improved assay and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. - PubMed
-
- Bordo D, Djinovic K, Bolognesi M. Conserved patterns in the Cu,Zn superoxide dismutase family. J Mol Biol. 1994;238:366–386. - PubMed
-
- Bowler C, van Montague M, Inzé D. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:83–116.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

