New class of small nonpeptidyl compounds blocks Plasmodium falciparum development in vitro by inhibiting plasmepsins
- PMID: 11502532
- PMCID: PMC90695
- DOI: 10.1128/AAC.45.9.2577-2584.2001
New class of small nonpeptidyl compounds blocks Plasmodium falciparum development in vitro by inhibiting plasmepsins
Abstract
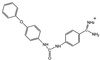
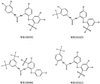
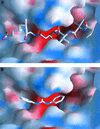
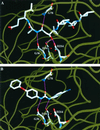
Malarial parasites rely on aspartic proteases called plasmepsins to digest hemoglobin during the intraerythrocytic stage. Plasmepsins from Plasmodium falciparum and Plasmodium vivax have been cloned and expressed for a variety of structural and enzymatic studies. Recombinant plasmepsins possess kinetic similarity to the native enzymes, indicating their suitability for target-based antimalarial drug development. We developed an automated assay of P. falciparum plasmepsin II and P. vivax plasmepsin to quickly screen compounds in the Walter Reed chemical database. A low-molecular-mass (346 Da) diphenylurea derivative (WR268961) was found to inhibit plasmepsins with a K(i) of 1 to 6 microM. This compound appears to be selective for plasmepsin, since it is a poor inhibitor of the human aspartic protease cathepsin D (K(i) greater than 280 microM). WR268961 inhibited the growth of P. falciparum strains W2 and D6, with 50% inhibitory concentrations ranging from 0.03 to 0.16 microg/ml, but was much less toxic to mammalian cells. The Walter Reed chemical database contains over 1,500 compounds with a diphenylurea core structure, 9 of which inhibit the plasmepsins, with K(i) values ranging from 0.05 to 0.68 microM. These nine compounds show specificity for the plasmepsins over human cathepsin D, but they are poor inhibitors of P. falciparum growth in vitro. Computational docking experiments indicate how diphenylurea compounds bind to the plasmepsin active site and inhibit the enzyme.
Figures



References
-
- Bailly E, Jambou R, Savel J, Jaureguiberry G. Plasmodium falciparum: differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and pepstatin A (aspartyl protease inhibitor) J Protozool. 1992;39:593–599. - PubMed
-
- Carroll C D, Orlowski M. Screening aspartyl proteases with combinatorial libraries. Adv Exp Med Biol. 1998;436:375–380. - PubMed
-
- Carroll C D, Johnson T O, Tao S, Lauri G, Orlowski M, Gluzman I Y, Goldberg D E, Dolle R E. Evaluation of a structure-based statine cyclic diamino amide encoded combinatorial library against plasmepsin II and cathepsin D. Bioorg Med Chem Lett. 1998;8:3203–3206. - PubMed
-
- Dame J B, Reddy G R, Yowell C A, Dunn B M, Kay J, Berry C. Sequence, expression and modeled structure of an aspartic proteinase from the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1994;64:177–190. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

