SOX6 binds CtBP2 to repress transcription from the Fgf-3 promoter
- PMID: 11504872
- PMCID: PMC55854
- DOI: 10.1093/nar/29.16.3347
SOX6 binds CtBP2 to repress transcription from the Fgf-3 promoter
Abstract
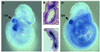
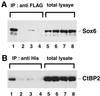
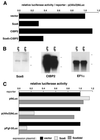
Fgf-3 is expressed in a complex pattern during mouse development. Previously, an essential regulatory element PS4A was identified in the promoter region, and shown to bind at least three factors. To identify the transcription factor(s), we used a yeast one-hybrid screen and obtained a novel Sox6 cDNA (SOX6D). When introduced into cells it strongly repressed activity from both an Fgf-3 reporter gene as well as an artificial promoter containing three PS4A elements. In situ hybridisation analysis showed that Sox6 and Fgf-3 are co-expressed in the otic vesicle of E9.5 mouse embryos in a mutually exclusive pattern, consistent with a repression of Fgf-3 transcription by SOX6. To characterise additional factor(s) involved in Fgf-3 gene repression, a yeast two-hybrid screen was used with the N-terminal portion of SOX6D. Mouse CtBP2 cDNA clones were isolated and shown to bind SOX6 in yeast and mammalian cells. Furthermore, mutational analysis of SOX6 showed that binding to CtBP2, and its responsiveness to this co-repressor, were dependent on a short amino acid sequence motif PLNLSS. Co-expression studies in NIH3T3 cells showed that SOX6 and CtBP2 co-operate to repress activity from the Fgf-3 promoter through the enhancer element PS4A. These results show that SOX6 can recruit CtBP2 to repress transcription from the Fgf-3 promoter.
Figures





References
-
- Ornitz D.M. (2000) FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays, 22, 108–112. - PubMed
-
- McKeehan W.L., Wang,F. and Kan,M. (1998) The heparan-sulfate fibroblast growth-factor family – diversity of structure and function. Prog. Nucleic Acids Res. Mol. Biol., 59, 135–176. - PubMed
-
- Yamaguchi T.P. and Rossant,J. (1995) Fibroblast growth-factors in mammalian development. Curr. Opin. Genet. Dev., 5, 485–491. - PubMed
-
- Martin G.R. (1998) The roles of FGFs in the early development of vertebrate limbs. Genes Dev., 12, 1571–1586. - PubMed
-
- Goldfarb M. (1996) Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev., 7, 311–325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

