The promoter of a lysosomal membrane transporter gene, CTNS, binds Sp-1, shares sequences with the promoter of an adjacent gene, CARKL, and causes cystinosis if mutated in a critical region
- PMID: 11505338
- PMCID: PMC1226058
- DOI: 10.1086/323484
The promoter of a lysosomal membrane transporter gene, CTNS, binds Sp-1, shares sequences with the promoter of an adjacent gene, CARKL, and causes cystinosis if mutated in a critical region
Abstract
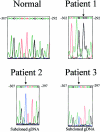
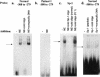
Although >55 CTNS mutations occur in patients with the lysosomal storage disorder cystinosis, no regulatory mutations have been reported, because the promoter has not been defined. Using CAT reporter constructs of sequences 5' to the CTNS coding sequence, we identified the CTNS promoter as the region encompassing nucleotides -316 to +1 with respect to the transcription start site. This region contains an Sp-1 regulatory element (GGCGGCG) at positions -299 to -293, which binds authentic Sp-1, as shown by electrophoretic-mobility-shift assays. Three patients exhibited mutations in the CTNS promoter. One patient with nephropathic cystinosis carried a -295 G-->C substitution disrupting the Sp-1 motif, whereas two patients with ocular cystinosis displayed a -303 G-->T substitution in one case and a -303 T insertion in the other case. Each mutation drastically reduced CAT activity when inserted into a reporter construct. Moreover, each failed either to cause a mobility shift when exposed to nuclear extract or to compete with the normal oligonucleotide's mobility shift. The CTNS promoter region shares 41 nucleotides with the promoter region of an adjacent gene of unknown function, CARKL, whose start site is 501 bp from the CTNS start site. However, the patients' CTNS promoter mutations have no effect on CARKL promoter activity. These findings suggest that the CTNS promoter region should be examined in patients with cystinosis who have fewer than two coding-sequence mutations.
Figures


References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CTNS [MIM 219800] and CARKL [MIM 605060])
References
-
- Ame J-C, Schreiber V, Fraulob V, Dolle P, de Murcia G, Niedergang CP (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. J Biol Chem 276:11092–11099 - PubMed
-
- Anikster Y, Lucero C, Guo J, Huizing M, Shotelersuk V, Bernardini I, McDowell G, Iwata F, Kaiser-Kupfer MI, Jaffe R, Thoene J, Schneider JA, Gahl WA (2000) Ocular, non-nephropathic cystinosis: clinical, biochemical and molecular correlations. Pediatr Res 47:17–23 - PubMed
-
- Anikster Y, Lucero C, Touchman JW, Huizing M, McDowell G, Shotelersuk V, Green ED, Gahl WA (1999a) Identification and detection of the common 65-kb deletion breakpoint in the nephropathic cystinosis gene (CTNS). Mol Genet Metab 66:111–116 - PubMed
-
- Anikster Y, Shotelersuk V, Gahl WA (1999b) CTNS mutations in patients with cystinosis. Hum Mutat 14:454–458 - PubMed
-
- Attard M, Jean G, Forestier L, Cherqui S, van’t Hoff W, Broyer M, Antignac C, Town M (1999) Severity of phenotype in cystinosis varies with mutations in the CTNS gene: predicted effect on the model of cystinosin. Hum Mol Genet 8:2507–2514 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

