Biochemical characterization of the helper component of Cauliflower mosaic virus
- PMID: 11507199
- PMCID: PMC115099
- DOI: 10.1128/jvi.75.18.8538-8546.2001
Biochemical characterization of the helper component of Cauliflower mosaic virus
Abstract
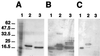
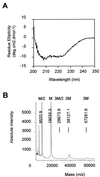
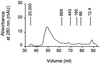
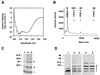
The helper component of Cauliflower mosaic virus is encoded by viral gene II. This protein (P2) is dispensable for virus replication but required for aphid transmission. The purification of P2 has never been reported, and hence its biochemical properties are largely unknown. We produced the P2 protein via a recombinant baculovirus with a His tag fused at the N terminus. The fusion protein was purified by affinity chromatography in a soluble and biologically active form. Matrix-assisted laser desorption time-of-flight mass spectrometry demonstrated that P2 is not posttranslationally modified. UV circular dichroism revealed the secondary structure of P2 to be 23% alpha-helical. Most alpha-helices are suggested to be located in the C-terminal domain. Using size exclusion chromatography and aphid transmission testing, we established that the active form of P2 assembles as a huge soluble oligomer containing 200 to 300 subunits. We further showed that P2 can also polymerize as long paracrystalline filaments. We mapped P2 domains involved in P2 self-interaction, presumably through coiled-coil structures, one of which is proposed to form a parallel trimer. These regions have previously been reported to also interact with viral P3, another protein involved in aphid transmission. Possible interference between the two types of interaction is discussed with regard to the biological activity of P2.
Figures



Similar articles
-
A coiled-coil interaction mediates cauliflower mosaic virus cell-to-cell movement.Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):6219-24. doi: 10.1073/pnas.0407731102. Epub 2005 Apr 18. Proc Natl Acad Sci U S A. 2005. PMID: 15837934 Free PMC article.
-
The N-terminus of the cauliflower mosaic virus aphid transmission protein P2 is involved in transmission body formation and microtubule interaction.Virus Res. 2021 May;297:198356. doi: 10.1016/j.virusres.2021.198356. Epub 2021 Mar 2. Virus Res. 2021. PMID: 33667624
-
Structural insights into the molecular mechanisms of cauliflower mosaic virus transmission by its insect vector.J Virol. 2010 May;84(9):4706-13. doi: 10.1128/JVI.02662-09. Epub 2010 Feb 24. J Virol. 2010. PMID: 20181714 Free PMC article.
-
The open reading frame III product of cauliflower mosaic virus forms a tetramer through a N-terminal coiled-coil.J Biol Chem. 1998 Oct 30;273(44):29015-21. doi: 10.1074/jbc.273.44.29015. J Biol Chem. 1998. PMID: 9786907
-
Plant pararetroviruses: interactions of cauliflower mosaic virus with plants and insects.Curr Opin Virol. 2013 Dec;3(6):629-38. doi: 10.1016/j.coviro.2013.08.014. Epub 2013 Sep 25. Curr Opin Virol. 2013. PMID: 24075119 Review.
Cited by
-
A coiled-coil interaction mediates cauliflower mosaic virus cell-to-cell movement.Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):6219-24. doi: 10.1073/pnas.0407731102. Epub 2005 Apr 18. Proc Natl Acad Sci U S A. 2005. PMID: 15837934 Free PMC article.
-
A single amino acid position in the helper component of cauliflower mosaic virus can change the spectrum of transmitting vector species.J Virol. 2005 Nov;79(21):13587-93. doi: 10.1128/JVI.79.21.13587-13593.2005. J Virol. 2005. PMID: 16227279 Free PMC article.
-
Plant virus transmission from the insect point of view.Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):17905-6. doi: 10.1073/pnas.0709178104. Epub 2007 Nov 7. Proc Natl Acad Sci U S A. 2007. PMID: 17989216 Free PMC article. No abstract available.
-
Identification of Plant Virus Receptor Candidates in the Stylets of Their Aphid Vectors.J Virol. 2018 Jun 29;92(14):e00432-18. doi: 10.1128/JVI.00432-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29769332 Free PMC article.
-
The cauliflower mosaic virus transmission helper protein P2 modifies directly the probing behavior of the aphid vector Myzus persicae to facilitate transmission.PLoS Pathog. 2023 Feb 6;19(2):e1011161. doi: 10.1371/journal.ppat.1011161. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36745680 Free PMC article.
References
-
- Armour S L, Melcher U, Pirone T P, Lyttle D J, Essenberg R C. Helper component for aphid transmission encoded by region II of cauliflower mosaic virus DNA. Virology. 1983;129:25–30. - PubMed
-
- Blanc S, Cerutti M, Chaabihi H, Louis C, Devauchelle G, Hull R. Gene II product of an aphid-nontransmissible isolate of cauliflower mosaic virus expressed in a baculovirus system possesses aphid transmission factor activity. Virology. 1993;192:651–654. - PubMed
-
- Blanc S, Cerutti M, Usmany M, Vlak J M, Hull R. Biological activity of cauliflower mosaic virus aphid transmission factor expressed in a heterologous system. Virology. 1993;192:643–650. - PubMed
-
- Blanc, S., E. Hébrard, M. Drucker, and R. Froissart. Molecular basis of vector transmission: caulimoviruses. In K. Harris, O. P. Smith, and J. E. Duffus (ed.), Virus-insect-plant interactions, in press. Academic Press, San Diego, Calif.
-
- Blanc S, Schmidt I, Kuhl G, Esperandieu P, Lebeurier G, Hull R, Cerutti M, Louis C. Paracrystalline structure of cauliflower mosaic virus aphid transmission factor produced both in plants and in a heterologous system and relationship with a solubilized active form. Virology. 1993;197:283–292. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

