Xenopus U3 snoRNA GAC-Box A' and Box A sequences play distinct functional roles in rRNA processing
- PMID: 11509664
- PMCID: PMC87338
- DOI: 10.1128/MCB.21.18.6210-6221.2001
Xenopus U3 snoRNA GAC-Box A' and Box A sequences play distinct functional roles in rRNA processing
Abstract
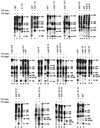
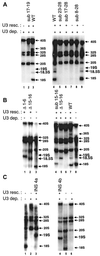
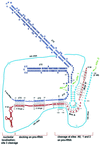
Mutations in the 5' portion of Xenopus U3 snoRNA were tested for function in oocytes. The results revealed a new cleavage site (A0) in the 3' region of vertebrate external transcribed spacer sequences. In addition, U3 mutagenesis uncoupled cleavage at sites 1 and 2, flanking the 5' and 3' ends of 18S rRNA, and generated novel intermediates: 19S and 18.5S pre-rRNAs. Furthermore, specific nucleotides in Xenopus U3 snoRNA that are required for cleavages in pre-rRNA were identified: box A is essential for site A0 cleavage, the GAC-box A' region is necessary for site 1 cleavage, and the 3' end of box A' and flanking nucleotides are required for site 2 cleavage. Differences between metazoan and yeast U3 snoRNA-mediated rRNA processing are enumerated. The data support a model where metazoan U3 snoRNA acts as a bridge to draw together the 5' and 3' ends of the 18S rRNA coding region within pre-rRNA to coordinate their cleavage.
Figures





References
-
- Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. - PubMed
-
- Ajuh P M, Heeney P A, Maden B E H. Xenopus borealis and Xenopus laevis 28S ribosomal DNA and the complete 40S ribosomal precursor RNA coding units of both species. Proc R Soc Lond B. 1991;245:65–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
