H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions
- PMID: 11509669
- PMCID: PMC87352
- DOI: 10.1128/MCB.21.18.6270-6279.2001
H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions
Abstract
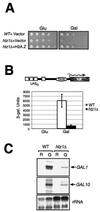
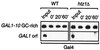
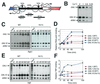
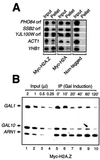
Evolutionarily conserved variant histone H2A.Z has been recently shown to regulate gene transcription in Saccharomyces cerevisiae. Here we show that loss of H2A.Z in this organism negatively affects the induction of GAL genes. Importantly, fusion of the H2A.Z C-terminal region to S phase H2A without its corresponding C-terminal region can mediate the variant histone's specialized function in GAL1-10 gene induction, and it restores the slow-growth phenotype of cells with a deletion of HTZ1. Furthermore, we show that the C-terminal region of H2A.Z can interact with some components of the transcriptional apparatus. In cells lacking H2A.Z, recruitment of RNA polymerase II and TATA-binding protein to the GAL1-10 promoters is significantly diminished under inducing conditions. Unexpectedly, we also find that H2A.Z is required to globally maintain chromatin integrity under GAL gene-inducing conditions. We hypothesize that H2A.Z can positively regulate gene transcription, at least in part, by modulating interactions with RNA polymerase II-associated factors at certain genes under specific cell growth conditions.
Figures


References
-
- Bash R, Lohr D. Yeast chromatin structure and regulation of GAL gene expression. Prog Nucleic Acid Res Mol Biol. 2000;65:197–259. - PubMed
-
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. - PubMed
-
- Clarkson M J, Wells J R E, Gibson F, Saint R, Tremethick D J. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. - PubMed
-
- Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle and developmentally regulated promoter. Cell. 1999;97:299–311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
