High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification
- PMID: 11511324
- PMCID: PMC37402
- DOI: 10.1186/1471-2164-2-4
High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification
Abstract
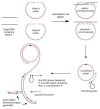
Background: Single nucleotide polymorphisms (SNPs) are the foundation of powerful complex trait and pharmacogenomic analyses. The availability of large SNP databases, however, has emphasized a need for inexpensive SNP genotyping methods of commensurate simplicity, robustness, and scalability. We describe a solution-based, microtiter plate method for SNP genotyping of human genomic DNA. The method is based upon allele discrimination by ligation of open circle probes followed by rolling circle amplification of the signal using fluorescent primers. Only the probe with a 3' base complementary to the SNP is circularized by ligation.
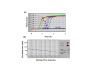
Results: SNP scoring by ligation was optimized to a 100,000 fold discrimination against probe mismatched to the SNP. The assay was used to genotype 10 SNPs from a set of 192 genomic DNA samples in a high-throughput format. Assay directly from genomic DNA eliminates the need to preamplify the target as done for many other genotyping methods. The sensitivity of the assay was demonstrated by genotyping from 1 ng of genomic DNA. We demonstrate that the assay can detect a single molecule of the circularized probe.
Conclusions: Compatibility with homogeneous formats and the ability to assay small amounts of genomic DNA meets the exacting requirements of automated, high-throughput SNP scoring.
Figures




References
-
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79839. - DOI - PubMed
-
- Landegren U, Nilsson M, Kwok PY. Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res. 1998;8:769–776. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

