Functional comparison of the nematode Hox gene lin-39 in C. elegans and P. pacificus reveals evolutionary conservation of protein function despite divergence of primary sequences
- PMID: 11511546
- PMCID: PMC312764
- DOI: 10.1101/gad.200601
Functional comparison of the nematode Hox gene lin-39 in C. elegans and P. pacificus reveals evolutionary conservation of protein function despite divergence of primary sequences
Abstract
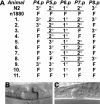
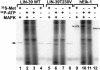
Hox transcription factors have been implicated in playing a central role in the evolution of animal morphology. Many studies indicate the evolutionary importance of regulatory changes in Hox genes, but little is known about the role of functional changes in Hox proteins. In the nematodes Pristionchus pacificus and Caenorhabditis elegans, developmental processes can be compared at the cellular, genetic, and molecular levels and differences in gene function can be identified. The Hox gene lin-39 is involved in the regulation of nematode vulva development. Comparison of known lin-39 mutations in P. pacificus and C. elegans revealed both conservation and changes of gene function. Here, we study evolutionary changes of lin-39 function using hybrid transgenes and site-directed mutagenesis in an in vivo assay using C. elegans lin-39 mutants. Our data show that despite the functional differences of LIN-39 between the two species, Ppa-LIN-39, when driven by Cel-lin-39 regulatory elements, can functionally replace Cel-lin-39. Furthermore, we show that the MAPK docking and phosphorylation motifs unique for Cel-LIN-39 are dispensable for Cel-lin-39 function. Therefore, the evolution of lin-39 function is driven by changes in regulatory elements rather than changes in the protein itself.
Figures




Similar articles
-
The pax-3 gene is involved in vulva formation in Pristionchus pacificus and is a target of the Hox gene lin-39.Development. 2007 Sep;134(17):3111-9. doi: 10.1242/dev.008375. Epub 2007 Jul 25. Development. 2007. PMID: 17652349
-
HAIRY-like transcription factors and the evolution of the nematode vulva equivalence group.Curr Biol. 2006 Jul 25;16(14):1386-94. doi: 10.1016/j.cub.2006.06.058. Curr Biol. 2006. PMID: 16860737
-
The adaptable lin-39.Nat Genet. 2001 Oct;29(2):106-7. doi: 10.1038/ng1001-106. Nat Genet. 2001. PMID: 11586287
-
A class act: conservation of homeodomain protein functions.Dev Suppl. 1994:61-77. Dev Suppl. 1994. PMID: 7579525 Review.
-
Evolution of the vertebrate Hox homeobox genes.Bioessays. 1992 Apr;14(4):245-52. doi: 10.1002/bies.950140408. Bioessays. 1992. PMID: 1350721 Review.
Cited by
-
Limited microsynteny between the genomes of Pristionchus pacificus and Caenorhabditis elegans.Nucleic Acids Res. 2003 May 15;31(10):2553-60. doi: 10.1093/nar/gkg359. Nucleic Acids Res. 2003. PMID: 12736304 Free PMC article.
-
Population genetics of Caenorhabditis elegans: the paradox of low polymorphism in a widespread species.Genetics. 2003 Jan;163(1):147-57. doi: 10.1093/genetics/163.1.147. Genetics. 2003. PMID: 12586703 Free PMC article.
-
Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene.Genes Dev. 2002 Sep 15;16(18):2345-9. doi: 10.1101/gad.996302. Genes Dev. 2002. PMID: 12231624 Free PMC article.
-
LIN-39/Hox triggers cell division and represses EFF-1/fusogen-dependent vulval cell fusion.Genes Dev. 2002 Dec 15;16(24):3136-41. doi: 10.1101/gad.251202. Genes Dev. 2002. PMID: 12502736 Free PMC article.
-
Hox proteins interact to pattern neuronal subtypes in Caenorhabditis elegans males.Genetics. 2022 Apr 4;220(4):iyac010. doi: 10.1093/genetics/iyac010. Genetics. 2022. PMID: 35137058 Free PMC article.
References
-
- Anderson P, Kimble J. mRNA and translation. In: Riddle DL, et al., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 185–208. - PubMed
-
- Beitel GJ, Tuck S, Greenwald I, Horvitz HR. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes & Dev. 1995;9:3149–3162. - PubMed
-
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. - PubMed
-
- Clandinin TR, Katz WS, Sternberg PW. Caenorhabditis elegans hom-C genes regulate the response of vulval precursor cells to inductive signal. Dev Biol. 1997;182:150–161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
