Hybrid protein between ribosomal protein S16 and RimM of Escherichia coli retains the ribosome maturation function of both proteins
- PMID: 11514519
- PMCID: PMC95418
- DOI: 10.1128/JB.183.18.5352-5357.2001
Hybrid protein between ribosomal protein S16 and RimM of Escherichia coli retains the ribosome maturation function of both proteins
Abstract
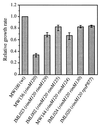
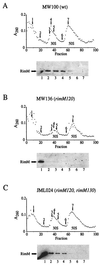
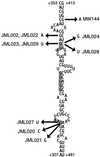
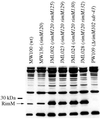
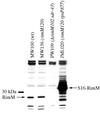
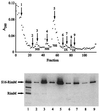
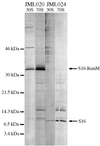
The RimM protein in Escherichia coli is associated with free 30S ribosomal subunits but not with 70S ribosomes and is important for efficient maturation of the 30S subunits. A mutant lacking RimM shows a sevenfold-reduced growth rate and a reduced translational efficiency. Here we show that a double alanine-for-tyrosine substitution in RimM prevents it from associating with the 30S subunits and reduces the growth rate of E. coli approximately threefold. Several faster-growing derivatives of the rimM amino acid substitution mutant were found that contain suppressor mutations which increased the amount of the RimM protein by two different mechanisms. Most of the suppressor mutations destabilized a secondary structure in the rimM mRNA, which previously was shown to decrease the synthesis of RimM by preventing the access of the ribosomes to the translation initiation region on the rimM mRNA. Three other independently isolated suppressor mutations created a fusion between rpsP, encoding the ribosomal protein S16, and rimM on the chromosome as a result of mutations in the rpsP stop codon preceding rimM. A severalfold-higher amount of the produced hybrid S16-RimM protein in the suppressor strains than of the native-sized RimM in the original substitution mutant seems to explain the suppression. The S16-RimM protein but not any native-size ribosomal protein S16 was found both in free 30S ribosomal subunits and in translationally active 70S ribosomes of the suppressor strains. This suggests that the hybrid protein can substitute for S16, which is an essential protein probably because of its role in ribosome assembly. Thus, the S16-RimM hybrid protein seems capable of carrying out the important functions that native S16 and RimM have in ribosome biogenesis.
Figures
Similar articles
-
The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits.RNA. 2004 Nov;10(11):1798-812. doi: 10.1261/rna.7720204. RNA. 2004. PMID: 15496525 Free PMC article.
-
RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli.J Bacteriol. 1998 Jan;180(1):73-82. doi: 10.1128/JB.180.1.73-82.1998. J Bacteriol. 1998. PMID: 9422595 Free PMC article.
-
A novel ribosome-associated protein is important for efficient translation in Escherichia coli.J Bacteriol. 1997 Jul;179(14):4567-74. doi: 10.1128/jb.179.14.4567-4574.1997. J Bacteriol. 1997. PMID: 9226267 Free PMC article.
-
Ribosome biogenesis and the translation process in Escherichia coli.Microbiol Mol Biol Rev. 2007 Sep;71(3):477-94. doi: 10.1128/MMBR.00013-07. Microbiol Mol Biol Rev. 2007. PMID: 17804668 Free PMC article. Review.
-
The Discovery of Ribosomal Protein bL31 from Escherichia coli: A Long Story Revisited.Int J Mol Sci. 2023 Feb 8;24(4):3445. doi: 10.3390/ijms24043445. Int J Mol Sci. 2023. PMID: 36834855 Free PMC article. Review.
Cited by
-
A coordinated proteomic approach for identifying proteins that interact with the E. coli ribosomal protein S12.J Proteome Res. 2013 Mar 1;12(3):1289-99. doi: 10.1021/pr3009435. Epub 2013 Feb 1. J Proteome Res. 2013. PMID: 23305560 Free PMC article.
-
Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation.J Mol Biol. 2005 May 27;349(1):47-59. doi: 10.1016/j.jmb.2005.03.075. Epub 2005 Apr 12. J Mol Biol. 2005. PMID: 15876367 Free PMC article.
-
The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits.RNA. 2004 Nov;10(11):1798-812. doi: 10.1261/rna.7720204. RNA. 2004. PMID: 15496525 Free PMC article.
-
Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement.RNA. 2013 Jun;19(6):789-802. doi: 10.1261/rna.037523.112. Epub 2013 Apr 23. RNA. 2013. PMID: 23611982 Free PMC article.
-
Ribosome Pool Engineering Increases Protein Biosynthesis Yields.ACS Cent Sci. 2024 Mar 20;10(4):871-881. doi: 10.1021/acscentsci.3c01413. eCollection 2024 Apr 24. ACS Cent Sci. 2024. PMID: 38680563 Free PMC article.
References
-
- Bonnefoy E. The ribosomal S16 protein of Escherichia coli displaying a DNA-nicking activity binds to cruciform DNA. Eur J Biochem. 1997;247:852–859. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

