The BPAG1 locus: Alternative splicing produces multiple isoforms with distinct cytoskeletal linker domains, including predominant isoforms in neurons and muscles
- PMID: 11514586
- PMCID: PMC2196450
- DOI: 10.1083/jcb.200012098
The BPAG1 locus: Alternative splicing produces multiple isoforms with distinct cytoskeletal linker domains, including predominant isoforms in neurons and muscles
Abstract
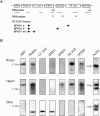
Bullous pemphigoid antigen 1 (BPAG1) is a member of the plakin family with cytoskeletal linker properties. Mutations in BPAG1 cause sensory neuron degeneration and skin fragility in mice. We have analyzed the BPAG1 locus in detail and found that it encodes different interaction domains that are combined in tissue-specific manners. These domains include an actin-binding domain (ABD), a plakin domain, a coiled coil (CC) rod domain, two different potential intermediate filament-binding domains (IFBDs), a spectrin repeat (SR)-containing rod domain, and a microtubule-binding domain (MTBD). There are at least three major forms of BPAG1: BPAG1-e (302 kD), BPAG1-a (615 kD), and BPAG1-b (834 kD). BPAG1-e has been described previously and consists of the plakin domain, the CC rod domain, and the first IFBD. It is the primary epidermal BPAG1 isoform, and its absence that is the likely cause of skin fragility in mutant mice. BPAG1-a is the major isoform in the nervous system and a homologue of the microtubule actin cross-linking factor, MACF. BPAG1-a is composed of the ABD, the plakin domain, the SR-containing rod domain, and the MTBD. The absence of BPAG1-a is the likely cause of sensory neurodegeneration in mutant mice. BPAG1-b is highly expressed in muscles, and has extra exons encoding a second IFBD between the plakin and SR-containing rod domains of BPAG1-a.
Figures
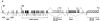
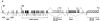
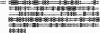

References
-
- Bernier, G., A. Brown, G. Dalpe, Y. De Repentigny, M. Mathieu, and R. Kothary. 1995. a. Dystonin expression in the developing nervous system predominates in the neurons that degenerate in dystonia musculorum mutant mice. Mol. Cell. Neurosci. 6:509–520. - PubMed
-
- Bernier, G., A. Brown, G. Dalpe, M. Mathieu, Y. De Repentigny, and R. Kothary. 1995. b. Dystonin transcripts are altered and their levels are reduced in the mouse neurological mutant dt24J. Biochem. Cell Biol. 73:605–609. - PubMed
-
- Bernier, G., Y. De Repentigny, M. Mathieu, S. David, and R. Kothary. 1998. Dystonin is an essential component of the Schwann cell cytoskeleton at the time of myelination. Development. 125:2135–2148. - PubMed
-
- Brown, A., G. Bernier, M. Mathieu, J. Rossant, and R. Kothary. 1995. The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat. Genet. 10:301–306. - PubMed
-
- Dalpe, G., N. Leclerc, A. Vallee, A. Messer, M. Mathieu, Y. De Repentigny, and R. Kothary. 1998. Dystonin is essential for maintaining neuronal cytoskeleton organization. Mol. Cell. Neurosci. 10:243–257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

