Biogenesis of Golgi stacks in imaginal discs of Drosophila melanogaster
- PMID: 11514618
- PMCID: PMC58596
- DOI: 10.1091/mbc.12.8.2308
Biogenesis of Golgi stacks in imaginal discs of Drosophila melanogaster
Abstract
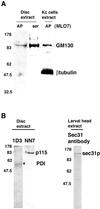
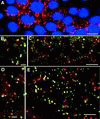
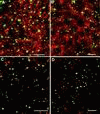
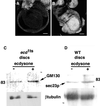
We provide a detailed description of Golgi stack biogenesis that takes place in vivo during one of the morphogenetic events in the lifespan of Drosophila melanogaster. In early third-instar larvae, small clusters consisting mostly of vesicles and tubules were present in epithelial imaginal disk cells. As larvae progressed through mid- and late-third instar, these larval clusters became larger but also increasingly formed cisternae, some of which were stacked. In white pupae, the typical Golgi stack was observed. We show that larval clusters are Golgi stack precursors by 1) localizing various Golgi-specific markers to the larval clusters by electron and immunofluorescence confocal microscopy, 2) driving this conversion in wild-type larvae incubated at 37 degrees C for 2 h, and 3) showing that this conversion does not take place in an NSF1 mutant (comt 17). The biological significance of this conversion became clear when we found that the steroid hormone 20-hydroxyecdysone (ecdysone) is critically involved in this conversion. In its absence, Golgi stack biogenesis did not occur and the larval clusters remained unaltered. We showed that dGM130 and sec23p expression increases approximately three- and fivefold, respectively, when discs are exposed to ecdysone in vivo and in vitro. Taken together, these results suggest that we have developed an in vivo system to study the ecdysone-triggered Golgi stack biogenesis.
Figures






Similar articles
-
Ecdysone triggers the expression of Golgi genes in Drosophila imaginal discs via broad-complex.Dev Biol. 2002 May 1;245(1):172-86. doi: 10.1006/dbio.2002.0632. Dev Biol. 2002. PMID: 11969264
-
Ecdysone signaling is required for proper organization and fluid secretion of stellate cells in the Malpighian tubules of Drosophila melanogaster.Int J Dev Biol. 2010;54(4):635-42. doi: 10.1387/ijdb.092910ng. Int J Dev Biol. 2010. PMID: 20209436
-
Reconstitution of vesiculated Golgi membranes into stacks of cisternae: requirement of NSF in stack formation.J Cell Biol. 1995 May;129(3):577-89. doi: 10.1083/jcb.129.3.577. J Cell Biol. 1995. PMID: 7730397 Free PMC article.
-
How many ways through the Golgi maze?Traffic. 2008 Mar;9(3):299-304. doi: 10.1111/j.1600-0854.2007.00690.x. Epub 2007 Dec 14. Traffic. 2008. PMID: 18088319 Review.
-
[Golgi units regulating diversity of glycosylation].Seikagaku. 2007 Jan;79(1):42-5. Seikagaku. 2007. PMID: 17319513 Review. Japanese. No abstract available.
Cited by
-
Drosophila myt1 is the major cdk1 inhibitory kinase for wing imaginal disc development.Genetics. 2008 Dec;180(4):2123-33. doi: 10.1534/genetics.108.093195. Epub 2008 Oct 20. Genetics. 2008. PMID: 18940789 Free PMC article.
-
Autophagic Removal of Farnesylated Carboxy-Terminal Lamin Peptides.Cells. 2018 Apr 23;7(4):33. doi: 10.3390/cells7040033. Cells. 2018. PMID: 29690642 Free PMC article.
-
Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin.Nature. 2009 Aug 20;460(7258):978-83. doi: 10.1038/nature08280. Epub 2009 Jul 26. Nature. 2009. PMID: 19633650
-
A distinct Golgi-targeting mechanism of dGM130 in Drosophila neurons.Front Mol Neurosci. 2023 Jun 2;16:1206219. doi: 10.3389/fnmol.2023.1206219. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37333614 Free PMC article.
-
Intracellular Golgi Complex organization reveals tissue specific polarity during zebrafish embryogenesis.Dev Dyn. 2016 Jun;245(6):678-91. doi: 10.1002/dvdy.24409. Epub 2016 May 3. Dev Dyn. 2016. PMID: 27043944 Free PMC article.
References
-
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. - PubMed
-
- Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. - PubMed
-
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

