Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways
- PMID: 11514630
- PMCID: PMC58608
- DOI: 10.1091/mbc.12.8.2482
Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways
Abstract
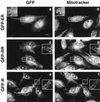
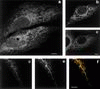
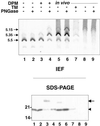
Many mitochondrial outer membrane (MOM) proteins have a transmembrane domain near the C terminus and an N-terminal cytosolic moiety. It is not clear how these tail-anchored (TA) proteins posttranslationally select their target, but C-terminal charged residues play an important role. To investigate how discrimination between MOM and endoplasmic reticulum (ER) occurs, we used mammalian cytochrome b(5), a TA protein existing in two, MOM or ER localized, versions. Substitution of the seven C-terminal residues of the ER isoform or of green fluorescent protein reporter constructs with one or two arginines resulted in MOM-targeted proteins, whereas a single C-terminal threonine caused promiscuous localization. To investigate whether targeting to MOM occurs from the cytosol or after transit through the ER, we tagged a MOM-directed construct with a C-terminal N-glycosylation sequence. Although in vitro this construct was efficiently glycosylated by microsomes, the protein expressed in vivo localized almost exclusively to MOM, and was nearly completely unglycosylated. The small fraction of glycosylated protein was in the ER and was not a precursor to the unglycosylated form. Thus, targeting occurs directly from the cytosol. Moreover, ER and MOM compete for the same polypeptide, explaining the dual localization of some TA proteins.
Figures







References
-
- Adamus G, Arendt A, Hargrave PA. Genetic control of antibody response to bovine rhodopsin in mice: epitope mapping of rhodopsin structure. J Neuroimmunol. 1991;34:89–97. - PubMed
-
- Akao Y, Otsuki YS, Kataoka S, Ito Y, Tsujimoto Y. Multiple subcellular localization of bcl-2: detection in nuclear outer membrane, endoplasmic reticulum, and mitochondrial membrane. Cancer Res. 1994;54:2468–2471. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1987.
-
- Bhakdi S, Weller U, Walev I, Martin E, Jonas D, Palmer M. A guide to the use of pore-forming toxins for controlled permeabilization of cell membranes. Med Microbiol Immunol. 1993;182:167–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

