Expression and phylogeny of claudins in vertebrate primordia
- PMID: 11517306
- PMCID: PMC56938
- DOI: 10.1073/pnas.171325898
Expression and phylogeny of claudins in vertebrate primordia
Abstract
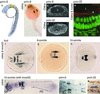
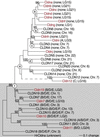
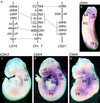
Claudins, the major transmembrane proteins of tight junctions, are members of the tetraspanin superfamily of proteins that mediate cellular adhesion and migration. Their functional importance is demonstrated by mutations in claudin genes that eliminate tight junctions in myelin and the testis, abolish Mg(2+) resorption in the kidney, and cause autosomal recessive deafness. Here we report that two paralogs among 15 claudin genes in the zebrafish, Danio rerio, are expressed in the otic and lateral-line placodes at their earliest stages of development. Related claudins in amphibians and mammals are expressed in a similar manner in vertebrate primordia such as sensory placodes, branchial arches, and limb buds. We also show that the claudin gene family may have expanded along the chordate stem lineage from urochordates to gnathostomes, in parallel with the elaboration of vertebrate characters. We propose that tight junctions not only form barriers in mature epithelia, but also participate in vertebrate morphogenesis.
Figures

Similar articles
-
Evolution of the vertebrate claudin gene family: insights from a basal vertebrate, the sea lamprey.Int J Dev Biol. 2016;60(1-3):39-51. doi: 10.1387/ijdb.150364nn. Int J Dev Biol. 2016. PMID: 27002805
-
Phylogenetic revision of the claudin gene family.Mar Genomics. 2013 Sep;11:17-26. doi: 10.1016/j.margen.2013.05.001. Epub 2013 May 28. Mar Genomics. 2013. PMID: 23726886
-
Evolution of developmental roles of Pax2/5/8 paralogs after independent duplication in urochordate and vertebrate lineages.BMC Biol. 2008 Aug 22;6:35. doi: 10.1186/1741-7007-6-35. BMC Biol. 2008. PMID: 18721460 Free PMC article.
-
Evolutionary origins of vertebrate placodes: insights from developmental studies and from comparisons with other deuterostomes.J Exp Zool B Mol Dev Evol. 2005 Jul 15;304(4):347-99. doi: 10.1002/jez.b.21055. J Exp Zool B Mol Dev Evol. 2005. PMID: 16003766 Review.
-
Molecular evidence from ascidians for the evolutionary origin of vertebrate cranial sensory placodes.J Exp Zool B Mol Dev Evol. 2005 Jul 15;304(4):340-6. doi: 10.1002/jez.b.21054. J Exp Zool B Mol Dev Evol. 2005. PMID: 15981200 Review.
Cited by
-
trpm7 regulation of in vivo cation homeostasis and kidney function involves stanniocalcin 1 and fgf23.Endocrinology. 2010 Dec;151(12):5700-9. doi: 10.1210/en.2010-0853. Epub 2010 Sep 29. Endocrinology. 2010. PMID: 20881241 Free PMC article.
-
Evidence for a role of tight junctions in regulating sodium permeability in zebrafish (Danio rerio) acclimated to ion-poor water.J Comp Physiol B. 2013 Feb;183(2):203-13. doi: 10.1007/s00360-012-0700-9. Epub 2012 Jul 29. J Comp Physiol B. 2013. PMID: 22843140
-
Transcriptional signature of progesterone in the fathead minnow ovary (Pimephales promelas).Gen Comp Endocrinol. 2013 Oct 1;192:159-69. doi: 10.1016/j.ygcen.2013.06.008. Epub 2013 Jun 22. Gen Comp Endocrinol. 2013. PMID: 23796460 Free PMC article.
-
Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes.Genome Res. 2004 Jul;14(7):1248-57. doi: 10.1101/gr.2400004. Epub 2004 Jun 14. Genome Res. 2004. PMID: 15197168 Free PMC article.
-
Claudin 13, a member of the claudin family regulated in mouse stress induced erythropoiesis.PLoS One. 2010 Sep 10;5(9):e12667. doi: 10.1371/journal.pone.0012667. PLoS One. 2010. PMID: 20844758 Free PMC article.
References
-
- Green C R, Bergquist P R. J Cell Sci. 1982;53:279–305.
-
- Tsukita S, Furuse M, Itoh M. Nat Rev Mol Cell Biol. 2001;2:285–293. - PubMed
-
- Lane N J. In: Tight Junctions. Cereijido M, editor. Boca Raton, FL: CRC; 1992. pp. 23–48.
-
- Bellen H J, Lu Y, Beckstead R, Bhat M A. Trends Neurosci. 1998;21:444–449. - PubMed
-
- Bronstein J M. Curr Opin Neurobiol. 2000;10:552–557. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

