Investigating stem cells in human colon by using methylation patterns
- PMID: 11517339
- PMCID: PMC58561
- DOI: 10.1073/pnas.191225998
Investigating stem cells in human colon by using methylation patterns
Abstract
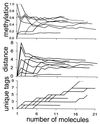
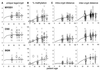
The stem cells that maintain human colon crypts are poorly characterized. To better determine stem cell numbers and how they divide, epigenetic patterns were used as cell fate markers. Methylation exhibits somatic inheritance and random changes that potentially record lifelong stem cell division histories as binary strings or tags in adjacent CpG sites. Methylation tag contents of individual crypts were sampled with bisulfite sequencing at three presumably neutral loci. Methylation increased with aging but varied between crypts and was mosaic within single crypts. Some crypts appeared to be quasi-clonal as they contained more unique tags than expected if crypts were maintained by single immortal stem cells. The complex epigenetic patterns were more consistent with a crypt niche model wherein multiple stem cells were present and replaced through periodic symmetric divisions. Methylation tags provide evidence that normal human crypts are long-lived, accumulate random methylation errors, and contain multiple stem cells that go through "bottlenecks" during life.
Figures




Comment in
-
Methylation patterns and mathematical models reveal dynamics of stem cell turnover in the human colon.Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10519-21. doi: 10.1073/pnas.201405498. Proc Natl Acad Sci U S A. 2001. PMID: 11553798 Free PMC article. No abstract available.
References
-
- Schmidt G H, Winton D J, Ponder B A J. Development (Cambridge, UK) 1988;103:785–790. - PubMed
-
- Potten C S, Loeffler M. Development (Cambridge, UK) 1990;110:1001–1020. - PubMed
-
- Bach S P, Renehan A G, Potten C S. Carcinogenesis. 2000;21:469–476. - PubMed
-
- Cairns J. Nature (London) 1975;255:197–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

