Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants
- PMID: 11517344
- PMCID: PMC56918
- DOI: 10.1073/pnas.191361598
Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants
Abstract
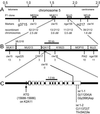
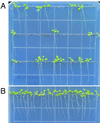
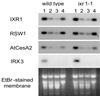
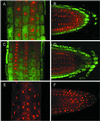
In many higher plants, cellulose synthesis is inhibited by isoxaben and thiazolidinone herbicides such as 5-tert-butyl-carbamoyloxy-3-(3-trifluromethyl) phenyl-4-thiazolidinone. Semidominant mutations at the IXR1 and IXR2 loci of Arabidopsis confer isoxaben and thiazolidinone resistance. Isolation of the IXR1 gene by map-based cloning revealed that it encodes the AtCESA3 isoform of cellulose synthase. The two known mutant alleles contain point mutations that replace glycine 998 with aspartic acid, and threonine 942 with isoleucine, respectively. The mutations occur in a highly conserved region of the enzyme near the carboxyl terminus that is well separated from the proposed active site. Although the IXR1 gene is expressed in the same cells as the structurally related RSW1 (AtCESA1) cellulose synthase gene, these two CESA genes are not functionally redundant.
Figures


References
-
- Lefebvre A, Maizonnier D, Gaudry J C, Clair D, Scalla R. Weed Res. 1987;27:125–134.
-
- Corio-Costet M-F, Agnese M D, Scalla R. Pestic Biochem Physiol. 1991;40:246–254.
-
- Heim D R, Skomp J R, Waldron C, Larrinua I M. Pestic Biochem Physiol. 1991;39:93–99.
-
- Corio-Costet M-F, Lherminier J, Scalla R. Pestic Biochem Physiol. 1991;40:255–265.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

