An integrated software suite for surface-based analyses of cerebral cortex
- PMID: 11522765
- PMCID: PMC131042
- DOI: 10.1136/jamia.2001.0080443
An integrated software suite for surface-based analyses of cerebral cortex
Abstract
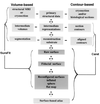
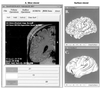
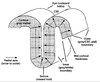
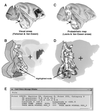
The authors describe and illustrate an integrated trio of software programs for carrying out surface-based analyses of cerebral cortex. The first component of this trio, SureFit (Surface Reconstruction by Filtering and Intensity Transformations), is used primarily for cortical segmentation, volume visualization, surface generation, and the mapping of functional neuroimaging data onto surfaces. The second component, Caret (Computerized Anatomical Reconstruction and Editing Tool Kit), provides a wide range of surface visualization and analysis options as well as capabilities for surface flattening, surface-based deformation, and other surface manipulations. The third component, SuMS (Surface Management System), is a database and associated user interface for surface-related data. It provides for efficient insertion, searching, and extraction of surface and volume data from the database.
Figures




Comment in
-
Human Brain Program Research Progress in biomedical imaging/neuroscience, 2001.J Am Med Inform Assoc. 2001 Sep-Oct;8(5):510-1. doi: 10.1136/jamia.2001.0080510. J Am Med Inform Assoc. 2001. PMID: 11522771 Free PMC article. No abstract available.
References
-
- Gattass R, Gross CG. Visual topography of striate projection zone (MT) in posterior superior temporal sulcus of the macaque. J Neurophysiol. 1981;46:621–38. - PubMed
-
- Van Essen DC, Drury HA, Anderson CH. An automated method for reconstructing complex surfaces, including the cerebral cortex. Soc Neurosci Abstr. 1999;25:1929.
-
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis, part I: Segmentation and surface reconstruction. NeuroImage. 1999;9:179–94. - PubMed
-
- Goebel R, Khorram-Sefat D, Muckli L, Hacker H, Singer W. Functional imaging of mirror and inverse reading reveals separate coactivated networks for oculomotion and spatial transformations. NeuroReport. 1998;9:713–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

