An enhancer element 6 kb upstream of the mouse HNF4alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors
- PMID: 11522818
- PMCID: PMC55877
- DOI: 10.1093/nar/29.17.3495
An enhancer element 6 kb upstream of the mouse HNF4alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors
Abstract
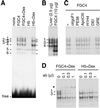
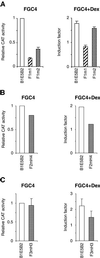
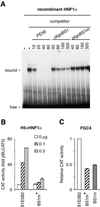
We have characterized a 700 bp enhancer element around -6 kb relative to the HNF4alpha1 transcription start. This element increases activity and confers glucocorticoid induction to a heterologous as well as the homologous promoters in differentiated hepatoma cells and is transactivated by HNF4alpha1, HNF4alpha7, HNF1alpha and HNF1beta in dedifferentiated hepatoma cells. A 240 bp sub-region conserves basal and hormone-induced enhancer activity. It contains HNF1, HNF4, HNF3 and C/EBP binding sites as shown by DNase I footprinting and electrophoretic mobility shift assays using nuclear extracts and/or recombinant HNF1alpha and HNF4alpha1. Mutation analyses showed that the HNF1 site is essential for HNF1alpha transactivation and is required for full basal enhancer activity, as is the C/EBP site. Glucocorticoid response element consensus sites which overlap the C/EBP, HNF4 and HNF3 sites are crucial for optimal hormonal induction. We present a model that accounts for weak expression of HNF4alpha1 in the embryonic liver and strong expression in the newborn/adult liver via the binding sites identified in the enhancer.
Figures





References
-
- Sladek F.M., Zhong,W., Lai,E. and Darnell,J.E.,Jr (1990) Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev., 4, 2353–2365. - PubMed
-
- Duncan S.A., Manova,K., Chen,W.S., Hoodless,P., Weinstein,D.C., Bachvarova,R.F. and Darnell,J.E.,Jr (1994) Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl Acad. Sci. USA, 91, 7598–7602. - PMC - PubMed
-
- Ang S.L. and Rossant,J. (1994) HNF-3β is essential for node and notochord formation in mouse development. Cell, 78, 561–574. - PubMed
-
- Barbacci E., Reber,M., Ott,M.O., Breillat,C., Huetz,F. and Cereghini,S. (1999) Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development, 126, 4795–4805. - PubMed
-
- Coffinier C., Thépot,D., Babinet,C., Yaniv,M. and Barra,J. (1999) Essential role for the homeoprotein vHNF1/HNF1β in visceral endoderm differentiation. Development, 126, 4785–4794. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

