Prediction of rho-independent transcriptional terminators in Escherichia coli
- PMID: 11522828
- PMCID: PMC55870
- DOI: 10.1093/nar/29.17.3583
Prediction of rho-independent transcriptional terminators in Escherichia coli
Abstract
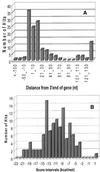
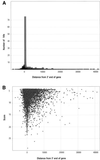
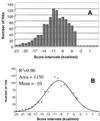
A new algorithm called RNAMotif containing RNA structure and sequence constraints and a thermodynamic scoring system was used to search for intrinsic rho-independent terminators in the Escherichia coli K-12 genome. We identified all 135 reported terminators and 940 putative terminator sequences beginning no more than 60 nt away from the 3'-end of the annotated transcription units (TU). Putative and reported terminators with the scores above our chosen threshold were found for 37 of the 53 non-coding RNA TU and for almost 50% of the 2592 annotated protein-encoding TU, which correlates well with the number of TU expected to contain rho-independent terminators. We also identified 439 terminators that could function in a bi-directional fashion, servicing one gene on the positive strand and a different gene on the negative strand. Approximately 700 additional termination signals in non-coding regions (NCR) far away from the nearest annotated gene were predicted. This number correlates well with the excess number of predicted 'orphan' promoters in the NCR, and these promoters and terminators may be associated with as yet unidentified TU. The significant number of high scoring hits that occurred within the reading frame of annotated genes suggests that either an additional component of rho-independent terminators exists or that a suppressive mechanism to prevent unwanted termination remains to be discovered.
Figures



References
-
- von Hippel P.H. (1998) An integrated model of the transcription complex in elongation, termination and editing. Science, 281, 660–665. - PubMed
-
- Reynolds R., Bermudez-Cruz,R.M. and Chamberlin,M.J. (1992) Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J. Mol. Biol., 224, 31–51. - PubMed
-
- von Hippel P.H. and Yager,T.D. (1992) The elongation-termination decision in transcription. Science, 255, 809–812. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

