Fluorogenic RT-PCR assay (TaqMan) for detection and classification of bovine viral diarrhea virus
- PMID: 11524161
- PMCID: PMC7117215
- DOI: 10.1016/s0378-1135(01)00390-x
Fluorogenic RT-PCR assay (TaqMan) for detection and classification of bovine viral diarrhea virus
Abstract
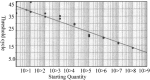
A single tube fluorogenic RT-PCR-based 'TaqMan' assay was developed for detection and classification of bovine viral diarrhea virus (BVDV). TaqMan-PCR was optimized to quantify BVD virus using the ABI PRISM 7700 sequence detection system and dual-labeled fluorogenic probes. Two different gene specific labeled fluorogenic probes for the 5' untranslated region (5' UTR) were used to differentiate between BVD types I and II. Sensitivity of the single tube TaqMan assay was compared with two-tube TaqMan assay and standard RT-PCR using 10-fold dilutions of RNA. Single tube TaqMan assay was 10-100-fold more sensitive than the two-tube TaqMan assay and the standardized single tube RT-PCR. Specificity of the assay was evaluated by testing different BVD virus strains and other bovine viruses. A total of 106 BVD positive and negative pooled or single serum samples, field isolates and reference strains were tested. Quantitation of cRNA from types I and II BVD virus was accomplished by a standard curve plotting cycle threshold values (C(T)) versus copy number. Single tube TaqMan-PCR assay was sensitive, specific and rapid for detection, quantitation and classification of BVD virus.
Figures

References
-
- Canal C.W, Hotzel I, de Almeida L.L, Michel Roehe P, Aoi M. Differentiation of classical swine fever virus from ruminant pestiviruses by reverse transcription and polymerase chain reaction. Vet. Microbiol. 1996;48:373–379. - PubMed
-
- Collet M.S, Larson R, Gold C, Strick D, Anderson D.K, Purchio A.F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. - PubMed
-
- Cortese, V.S., 1999. Food animal practice. Curr. Vet. Ther. 286–290.
-
- Drew T.W, Fenella Y, Pato D.J. The detection of bovine viral diarrhea virus in bulk milk samples by the use of a single-tube RT–PCR. Vet. Microbiol. 1999;64:145–154. - PubMed
-
- El-Kholy A.A, Bolin S.R, Ridpath J.F, Arab R.M.H, Abou-Zeid A.A, Hammam H.M, Platt K.B. Use of polymerase chain reaction to simultaneously detect and type bovine viral diarrhea viruses isolated from clinical specimens. Rev. Sci. Tech. Off. Int. Epiz. 1998;17(3):733–742. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

