nifH sequences and nitrogen fixation in type I and type II methanotrophs
- PMID: 11525998
- PMCID: PMC93122
- DOI: 10.1128/AEM.67.9.4009-4016.2001
nifH sequences and nitrogen fixation in type I and type II methanotrophs
Abstract
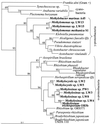
Some methane-oxidizing bacteria (methanotrophs) are known to be capable of expressing nitrogenase and utilizing N2 as a nitrogen source. However, no sequences are available for nif genes in these strains, and the known nitrogen-fixing methanotrophs are confined mainly to a few genera. The purpose of this work was to assess the nitrogen-fixing capabilities of a variety of methanotroph strains. nifH gene fragments from four type I methanotrophs and seven type II methanotrophs were PCR amplified and sequenced. Nitrogenase activity was confirmed in selected type I and type II strains by acetylene reduction. Activities ranged from 0.4 to 3.3 nmol/min/mg of protein. Sequence analysis shows that the nifH sequences from the type I and type II strains cluster with nifH sequences from other gamma proteobacteria and alpha proteobacteria, respectively. The translated nifH sequences from three Methylomonas strains show high identity (95 to 99%) to several published translated environmental nifH sequences PCR amplified from rice roots and a freshwater lake. The translated nifH sequences from the type II strains show high identity (94 to 99%) to published translated nifH sequences from a variety of environments, including rice roots, a freshwater lake, an oligotrophic ocean, and forest soil. These results provide evidence for nitrogen fixation in a broad range of methanotrophs and suggest that nitrogen-fixing methanotrophs may be widespread and important in the nitrogen cycling of many environments.
Figures

References
-
- Anthony C. The biochemistry of methylotrophs. London, England: Academic Press; 1982.
-
- Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methantrophs. Int J Syst Evol Bacteriol. 1993;43:735–753.
-
- Chu K-H, Alvarez-Cohen L. Trichloroethylene degradation by methane-oxidizing cultures grown with various nitrogen sources. Water Environ Res. 1996;68:76–82.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

