Obligate sulfide-dependent degradation of methoxylated aromatic compounds and formation of methanethiol and dimethyl sulfide by a freshwater sediment isolate, Parasporobacterium paucivorans gen. nov., sp. nov
- PMID: 11525999
- PMCID: PMC93123
- DOI: 10.1128/AEM.67.9.4017-4023.2001
Obligate sulfide-dependent degradation of methoxylated aromatic compounds and formation of methanethiol and dimethyl sulfide by a freshwater sediment isolate, Parasporobacterium paucivorans gen. nov., sp. nov
Abstract
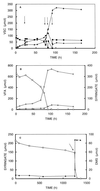
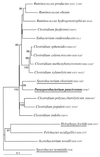
Methanethiol (MT) and dimethyl sulfide (DMS) have been shown to be the dominant volatile organic sulfur compounds in freshwater sediments. Previous research demonstrated that in these habitats MT and DMS are derived mainly from the methylation of sulfide. In order to identify the microorganisms that are responsible for this type of MT and DMS formation, several sulfide-rich freshwater sediments were amended with two potential methyl group-donating compounds, syringate and 3,4,5-trimethoxybenzoate (0.5 mM). The addition of these methoxylated aromatic compounds resulted in excess accumulation of MT and DMS in all sediment slurries even though methanogenic consumption of MT and DMS occurred. From one of the sediment slurries tested, a novel anaerobic bacterium was isolated with syringate as the sole carbon source. The strain, designated Parasporobacterium paucivorans, produced MT and DMS from the methoxy groups of syringate. The hydroxylated aromatic residue (gallate) was converted to acetate and butyrate. Like Sporobacterium olearium, another methoxylated aromatic compound-degrading bacterium, the isolate is a member of the XIVa cluster of the low-GC-content Clostridiales group. However, the new isolate differs from all other known methoxylated aromatic compound-degrading bacteria because it was able to degrade syringate in significant amounts only in the presence of sulfide.
Figures



References
-
- Achenbach L, Woese C. 16S and 23S rRNA-like primers. In: Robb F T, Place A R, Sowers K R, Scheier H J, DasSarma S, Fleischmann E M, editors. Archaea, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 521–523.
-
- Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981;130:255–261.
-
- Bak F, Finster K, Rothfuss F. Formation of dimethyl sulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol. 1992;157:529–534.
-
- Cazemier A E, Op den Camp H J M, Hackstein J H P, Vogels G D. Fibre digestion in arthropods. Comp Biochem Physiol. 1997;118A:101–109.
-
- Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

