Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling
- PMID: 11526021
- PMCID: PMC93145
- DOI: 10.1128/AEM.67.9.4177-4185.2001
Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling
Abstract
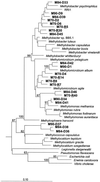
The diversity of methanotrophic bacteria associated with roots of submerged rice plants was assessed using cultivation-independent techniques. The research focused mainly on the retrieval of pmoA, which encodes the alpha subunit of the particulate methane monooxygenase. A novel methanotroph-specific community-profiling method was established using the terminal restriction fragment length polymorphism (T-RFLP) technique. The T-RFLP profiles clearly revealed a more complex root-associated methanotrophic community than did banding patterns obtained by pmoA-based denaturing gradient gel electrophoresis. The comparison of pmoA-based T-RFLP profiles obtained from rice roots and bulk soil of flooded rice microcosms suggested that there was a substantially higher abundance of type I methanotrophs on rice roots than in the bulk soil. These were affiliated to the genera Methylomonas, Methylobacter, Methylococcus, and to a novel type I methanotroph sublineage. By contrast, type II methanotrophs of the Methylocystis-Methylosinus group could be detected with high relative signal intensity in both soil and root compartments. Phylogenetic treeing analyses and a set of substrate-diagnostic amino acid residues provided evidence that a novel pmoA lineage was detected. This branched distinctly from all currently known methanotrophs. To examine whether the retrieval of pmoA provided a complete view of root-associated methanotroph diversity, we also assessed the diversity detectable by recovery of genes coding for subunits of soluble methane monooxygenase (mmoX) and methanol dehydrogenase (mxaF). In addition, both 16S rRNA and 16S ribosomal DNA (rDNA) were retrieved using a PCR primer set specific to type I methanotrophs. The overall methanotroph diversity detected by recovery of mmoX, mxaF, and 16S rRNA and 16S rDNA corresponded well to the diversity detectable by retrieval of pmoA.
Figures




Similar articles
-
Diversity of the particulate methane monooxygenase gene in methanotrophic samples from different rice field soils in China and the Philippines.Syst Appl Microbiol. 2002 Aug;25(2):267-74. doi: 10.1078/0723-2020-00104. Syst Appl Microbiol. 2002. PMID: 12353882
-
Diversity of methanotroph communities in a basalt aquifer.FEMS Microbiol Ecol. 2004 Jun 1;48(3):333-44. doi: 10.1016/j.femsec.2004.02.001. FEMS Microbiol Ecol. 2004. PMID: 19712303
-
Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms.Appl Environ Microbiol. 2001 Feb;67(2):623-31. doi: 10.1128/AEM.67.2.623-631.2001. Appl Environ Microbiol. 2001. PMID: 11157225 Free PMC article.
-
Diversity and Habitat Preferences of Cultivated and Uncultivated Aerobic Methanotrophic Bacteria Evaluated Based on pmoA as Molecular Marker.Front Microbiol. 2015 Dec 15;6:1346. doi: 10.3389/fmicb.2015.01346. eCollection 2015. Front Microbiol. 2015. PMID: 26696968 Free PMC article. Review.
-
Roses by other names: taxonomy of the Rhizobiaceae.J Bacteriol. 2003 May;185(10):2975-9. doi: 10.1128/JB.185.10.2975-2979.2003. J Bacteriol. 2003. PMID: 12730155 Free PMC article. Review. No abstract available.
Cited by
-
Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil.Appl Environ Microbiol. 2005 Mar;71(3):1671-3. doi: 10.1128/AEM.71.3.1671-1673.2005. Appl Environ Microbiol. 2005. PMID: 15746378 Free PMC article.
-
Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils.Appl Environ Microbiol. 2006 Feb;72(2):1346-54. doi: 10.1128/AEM.72.2.1346-1354.2006. Appl Environ Microbiol. 2006. PMID: 16461686 Free PMC article.
-
Diversity of oxygenase genes from methane- and ammonia-oxidizing bacteria in the Eastern Snake River Plain aquifer.Appl Environ Microbiol. 2005 Apr;71(4):2016-25. doi: 10.1128/AEM.71.4.2016-2025.2005. Appl Environ Microbiol. 2005. PMID: 15812034 Free PMC article.
-
Genotypic microbial community profiling: a critical technical review.Microb Ecol. 2007 Aug;54(2):276-89. doi: 10.1007/s00248-006-9199-5. Epub 2007 Mar 8. Microb Ecol. 2007. PMID: 17345133 Review.
-
Impact of plants on the diversity and activity of methylotrophs in soil.Microbiome. 2020 Mar 10;8(1):31. doi: 10.1186/s40168-020-00801-4. Microbiome. 2020. PMID: 32156318 Free PMC article.
References
-
- Amaral J A, Knowles R. Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett. 1995;126:215–220.
-
- Bodelier P L E, Roslev P, Henckel T, Frenzel P. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature. 2000;403:421–424. - PubMed
-
- Bodrossy L, Holmes E M, Holmes A J, Kovács K L, Murrell J C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol. 1997;168:493–503. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

