Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR
- PMID: 11526135
- PMCID: PMC88303
- DOI: 10.1128/JCM.39.9.3099-3103.2001
Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR
Abstract
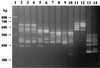
The emergence of coagulase-negative staphylococci not only as human pathogens but also as reservoirs of antibiotic resistance determinants requires the deployment and development of methods for their rapid and reliable identification. Internal transcribed spacer-PCR (ITS-PCR) was used to identify a collection of 617 clinical staphylococcal isolates. The amplicons were resolved in high-resolution agarose gels and visually compared with the patterns obtained for the control strains of 29 staphylococcal species. Of the 617 isolates studied, 592 (95.95%) were identified by ITS-PCR and included 11 species: 302 isolates of Staphylococcus epidermidis, 157 of S. haemolyticus, 79 of S. aureus, 21 of S. hominis, 14 of S. saprophyticus, 8 of S. warneri, 6 of S. simulans, 2 of S. lugdunensis, and 1 each of S. caprae, S. carnosus, and S. cohnii. All species analyzed had unique ITS-PCR patterns, although some were very similar, namely, the group S. saprophyticus, S. cohnii, S. gallinarum, S. xylosus, S. lentus, S. equorum, and S. chromogenes, the pair S. schleiferi and S. vitulus, and the pair S. piscifermentans and S. carnosus. Four species, S. aureus, S. caprae, S. haemolyticus, and S. lugdunensis, showed polymorphisms on their ITS-PCR patterns. ITS-PCR proved to be a valuable alternative for the identification of staphylococci, offering, within the same response time and at lower cost, higher reliability than the currently available commercial systems.
Figures


References
-
- Aires de Sousa M, Santos Sanches I, van Belkum A, van Leeuwen W, Verbrugh H, de Lencastre H. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping methods. Microb Drug Res. 1996;2:331–341. - PubMed
-
- Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:51–56. - PubMed
-
- Bes M, Guerin-Faublee V, Meugnier H, Etienne J, Freney J. Improvement of the identification of staphylococci isolated from bovine mammary infections using molecular methods. Vet Microbiol. 2000;71:287–294. - PubMed
-
- Dolzani L, Tonin E, Lagatolla C, Monti-Bragadin C. Typing of Staphylococcus aureus by amplification of the 16S–23S rRNA intergenic spacer sequences. FEMS Microbiol Lett. 1994;119:167–173. - PubMed
-
- Forsman P, Tilsala-Timisjärvi A, Alatossava T. Identification of staphylococcal and streptpcoccal causes of bovine mastitis using 16S–23S rRNA spacer regions. Microbiology. 1997;143:3491–3500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

