Multiple conformations of PEVK proteins detected by single-molecule techniques
- PMID: 11526214
- PMCID: PMC58526
- DOI: 10.1073/pnas.191189098
Multiple conformations of PEVK proteins detected by single-molecule techniques
Abstract
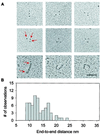
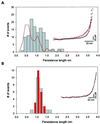
An important component of muscle elasticity is the PEVK region of titin, so named because of the preponderance of these amino acids. However, the PEVK region, similar to other elastomeric proteins, is thought to form a random coil and therefore its structure cannot be determined by standard techniques. Here we combine single-molecule electron microscopy and atomic force microscopy to examine the conformations of the human cardiac titin PEVK region. In contrast to a simple random coil, we have found that cardiac PEVK shows a wide range of elastic conformations with end-to-end distances ranging from 9 to 24 nm and persistence lengths from 0.4 to 2.5 nm. Individual PEVK molecules retained their distinctive elastic conformations through many stretch-relaxation cycles, consistent with the view that these PEVK conformers cannot be interconverted by force. The multiple elastic conformations of cardiac PEVK may result from varying degrees of proline isomerization. The single-molecule techniques demonstrated here may help elucidate the conformation of other proteins that lack a well-defined structure.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

