AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations
- PMID: 11526243
- PMCID: PMC56972
- DOI: 10.1073/pnas.171321298
AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations
Abstract
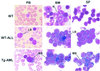
The t(8;21) is one of the most frequent chromosomal abnormalities associated with acute myeloid leukemia (AML). The translocation, which involves the AML1 gene on chromosome 21 and the ETO gene on chromosome 8, generates an AML1-ETO fusion transcription factor. To examine the effect of the AML1-ETO fusion protein on leukemogenesis, we made transgenic mice in which expression of AML1-ETO is under the control of the human MRP8 promoter (hMRP8-AML1-ETO). AML1-ETO is specifically expressed in myeloid cells, including common myeloid progenitors of hMRP8-AML1-ETO transgenic mice. The transgenic mice were healthy during their life spans, suggesting that AML1-ETO alone is not sufficient for leukemogenesis. However, after treatment of newborn hMRP8-AML1-ETO transgenic mice and their wild-type littermates with a strong DNA-alkylating mutagen, N-ethyl-N-nitrosourea, 55% of transgenic mice developed AML and the other 45% of transgenic mice and all of the wild-type littermates developed acute T lymphoblastic leukemia. Our results provide direct evidence that AML1-ETO is critical for causing myeloid leukemia, but one or more additional mutations are required for leukemogenesis. The hMRP8-AML1-ETO-transgenic mice provide an excellent model that can be used to isolate additional genetic events and to further understand the molecular pathogenesis of AML1-ETO-related leukemia.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

