Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli
- PMID: 11528004
- PMCID: PMC99035
- DOI: 10.1128/MMBR.65.3.422-444.2001
Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli
Abstract
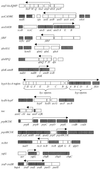
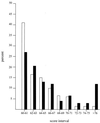
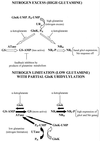
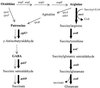
Sigma(54) has several features that distinguish it from other sigma factors in Escherichia coli: it is not homologous to other sigma subunits, sigma(54)-dependent expression absolutely requires an activator, and the activator binding sites can be far from the transcription start site. A rationale for these properties has not been readily apparent, in part because of an inability to assign a common physiological function for sigma(54)-dependent genes. Surveys of sigma(54)-dependent genes from a variety of organisms suggest that the products of these genes are often involved in nitrogen assimilation; however, many are not. Such broad surveys inevitably remove the sigma(54)-dependent genes from a potentially coherent metabolic context. To address this concern, we consider the function and metabolic context of sigma(54)-dependent genes primarily from a single organism, Escherichia coli, in which a reasonably complete list of sigma(54)-dependent genes has been identified by computer analysis combined with a DNA microarray analysis of nitrogen limitation-induced genes. E. coli appears to have approximately 30 sigma(54)-dependent operons, and about half are involved in nitrogen assimilation and metabolism. A possible physiological relationship between sigma(54)-dependent genes may be based on the fact that nitrogen assimilation consumes energy and intermediates of central metabolism. The products of the sigma(54)-dependent genes that are not involved in nitrogen metabolism may prevent depletion of metabolites and energy resources in certain environments or partially neutralize adverse conditions. Such a relationship may limit the number of physiological themes of sigma(54)-dependent genes within a single organism and may partially account for the unique features of sigma(54) and sigma(54)-dependent gene expression.
Figures

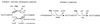
References
-
- Abouhamad W N, Manson M, Gibson M M, Higgins C F. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol Microbiol. 1991;5:1035–1047. - PubMed
-
- Adler S P, Purich D, Stadtman E R. Cascade control of Escherichia coli glutamine synthetase. Properties of the PII regulatory protein and the uridylyltransferase-uridylyl-removing enzyme. J Biol Chem. 1975;250:6264–6272. - PubMed
-
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

