Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis
- PMID: 11532947
- PMCID: PMC125267
- DOI: 10.1093/emboj/20.17.4836
Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis
Abstract
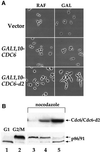
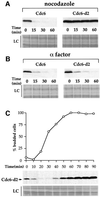
The Cdc6 DNA replication initiation factor is targeted for ubiquitin-mediated proteolysis by the E3 ubiquitin ligase SCF(CDC4) from the end of G1phase until mitosis in the budding yeast Saccharomyces cerevisiae. Here we describe a dominant-negative CDC6 mutant that, when overexpressed, arrests the cell cycle by inhibiting cyclin-dependent kinases (CDKs) and, thus, prevents passage through mitosis. This mutant protein inhibits CDKs more efficiently than wild-type Cdc6, in part because it is completely refractory to SCF(CDC4)-mediated proteolysis late in the cell cycle and consequently accumulates to high levels. The mutation responsible for this phenotype destroys a putative CDK phosphorylation site near the middle of the Cdc6 primary amino acid sequence. We show that this site lies within a novel Cdc4-interacting domain distinct from a Cdc4-interacting site identified previously near the N-terminus of the protein. We show that both sites can target Cdc6 for proteolysis in late G1/early S phase whilst only the newly identified site can target Cdc6 for proteolysis during mitosis.
Figures




Similar articles
-
The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast.EMBO J. 1997 Oct 1;16(19):5966-76. doi: 10.1093/emboj/16.19.5966. EMBO J. 1997. PMID: 9312054 Free PMC article.
-
Cdc6 degradation requires phosphodegron created by GSK-3 and Cdk1 for SCFCdc4 recognition in Saccharomyces cerevisiae.Mol Biol Cell. 2015 Jul 15;26(14):2609-19. doi: 10.1091/mbc.E14-07-1213. Epub 2015 May 20. Mol Biol Cell. 2015. PMID: 25995377 Free PMC article.
-
Activation of S-phase-promoting CDKs in late G1 defines a "point of no return" after which Cdc6 synthesis cannot promote DNA replication in yeast.Genes Dev. 1996 Jun 15;10(12):1516-31. doi: 10.1101/gad.10.12.1516. Genes Dev. 1996. PMID: 8666235
-
Multisite phosphorylation and the countdown to S phase.Cell. 2001 Dec 28;107(7):819-22. doi: 10.1016/s0092-8674(01)00620-1. Cell. 2001. PMID: 11779457 Review.
-
Redundant targeting of Isr1 by two CDKs in mitotic cells.Curr Genet. 2021 Feb;67(1):79-83. doi: 10.1007/s00294-020-01110-x. Epub 2020 Oct 15. Curr Genet. 2021. PMID: 33063175 Free PMC article. Review.
Cited by
-
The budding yeast GSK-3 homologue Mck1 is an essential component of the spindle position checkpoint.Open Biol. 2022 Nov;12(11):220203. doi: 10.1098/rsob.220203. Epub 2022 Nov 2. Open Biol. 2022. PMID: 36321416 Free PMC article.
-
The Hect domain E3 ligase Tom1 and the F-box protein Dia2 control Cdc6 degradation in G1 phase.J Biol Chem. 2012 Dec 28;287(53):44212-20. doi: 10.1074/jbc.M112.401778. Epub 2012 Nov 5. J Biol Chem. 2012. PMID: 23129771 Free PMC article.
-
Replication origins and timing of temporal replication in budding yeast: how to solve the conundrum?Curr Genomics. 2010 May;11(3):199-211. doi: 10.2174/138920210791110942. Curr Genomics. 2010. PMID: 21037857 Free PMC article.
-
Cdc6 is sequentially regulated by PP2A-Cdc55, Cdc14, and Sic1 for origin licensing in S. cerevisiae.Elife. 2022 Feb 10;11:e74437. doi: 10.7554/eLife.74437. Elife. 2022. PMID: 35142288 Free PMC article.
-
Functional dissection of the catalytic carboxyl-terminal domain of origin recognition complex subunit 1 (PfORC1) of the human malaria parasite Plasmodium falciparum.Eukaryot Cell. 2009 Sep;8(9):1341-51. doi: 10.1128/EC.00170-09. Epub 2009 Jul 24. Eukaryot Cell. 2009. PMID: 19633266 Free PMC article.
References
-
- Amon A., Tyers,M., Futcher,B. and Nasmyth,K. (1993) Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell, 74, 993–1007. - PubMed
-
- Calzada A., Sanchez,M., Sanchez,E. and Bueno,A. (2000) The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem., 275, 9734–9741. - PubMed
-
- Christianson T.W., Sikorski,R.S., Dante,M., Shero,J.H. and Hieter,P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene, 110, 119–122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

