Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants
- PMID: 11533152
- PMCID: PMC114457
- DOI: 10.1128/JVI.75.19.8888-8898.2001
Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants
Abstract
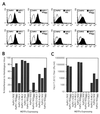
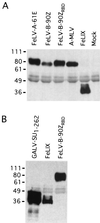
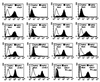
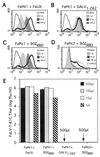
Cytopathic, T-cell-tropic feline leukemia viruses (FeLV-T) evolve from FeLV-A in infected animals and demonstrate host cell specificities that are distinct from those of their parent viruses. We recently identified two cellular proteins, FeLIX and Pit1, required for productive infection by these immunodeficiency-inducing FeLV-T variants (M. M. Anderson, A. S. Lauring, C. C. Burns, and J. Overbaugh, Science 287:1828-1830, 2000). FeLV-T is the first example of a naturally occurring type C retrovirus that requires two proteins to gain entry into target cells. FeLIX is an endogenous protein that is highly related to the N-terminal portion of the FeLV envelope protein, which includes the receptor-binding domain. Pit1 is a multiple-transmembrane phosphate transport protein that also functions as a receptor for FeLV-B. The FeLV-B envelope gene is derived by recombination with endogenous FeLV-like sequences, and its product can functionally substitute for FeLIX in facilitating entry through the Pit1 receptor. In the present study, we tested other retrovirus envelope surface units (SUs) with their cognate receptors to determine whether they also could mediate infection by FeLV-T. Cells were engineered to coexpress the transmembrane form of the envelope proteins and their cognate receptors, or SU protein was added as a soluble protein to cells expressing the receptor. Of the FeLV, murine leukemia virus, and gibbon ape leukemia virus envelopes tested, we found that only those with receptor-binding domains derived from endogenous FeLV could render cells permissive for FeLV-T. We also found that there is a strong preference for Pit1 as the transmembrane receptor. Specifically, FeLV-B SUs could efficiently mediate infection of cells expressing the Pit1 receptor but could only inefficiently mediate infection of cells expressing the Pit2 receptor, even though these SUs are able to bind to Pit2. Expression analysis of feline Pit1 and FeLIX suggests that FeLIX is likely the primary determinant of FeLV-T tropism. These results are discussed in terms of current models for retrovirus entry and the interrelationship among FeLV variants that evolve in vivo.
Figures


Similar articles
-
Genetic and biochemical analyses of receptor and cofactor determinants for T-cell-tropic feline leukemia virus infection.J Virol. 2002 Aug;76(16):8069-78. doi: 10.1128/jvi.76.16.8069-8078.2002. J Virol. 2002. PMID: 12134012 Free PMC article.
-
Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2.J Virol. 2001 Aug;75(15):6841-9. doi: 10.1128/JVI.75.15.6841-6849.2001. J Virol. 2001. PMID: 11435563 Free PMC article.
-
Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses.J Virol. 2001 Nov;75(22):10563-72. doi: 10.1128/JVI.75.22.10563-10572.2001. J Virol. 2001. PMID: 11602698 Free PMC article.
-
Viral genetic variation, AIDS, and the multistep nature of carcinogenesis: the feline leukemia virus model.Leukemia. 1996 Dec;10(12):1867-9. Leukemia. 1996. PMID: 8946923 Review.
-
Quantification of endogenous and exogenous feline leukemia virus sequences by real-time PCR assays.Vet Immunol Immunopathol. 2008 May 15;123(1-2):129-33. doi: 10.1016/j.vetimm.2008.01.027. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18295344 Review.
Cited by
-
Genetic and biochemical analyses of receptor and cofactor determinants for T-cell-tropic feline leukemia virus infection.J Virol. 2002 Aug;76(16):8069-78. doi: 10.1128/jvi.76.16.8069-8078.2002. J Virol. 2002. PMID: 12134012 Free PMC article.
-
Characterization of ferret Pit1 as a receptor of feline leukemia virus subgroup B.J Vet Med Sci. 2023 Mar 1;85(3):326-328. doi: 10.1292/jvms.22-0526. Epub 2023 Jan 19. J Vet Med Sci. 2023. PMID: 36653146 Free PMC article.
-
Importance of receptor usage, Fli1 activation, and mouse strain for the stem cell specificity of 10A1 murine leukemia virus leukemogenicity.J Virol. 2007 Jan;81(2):732-42. doi: 10.1128/JVI.01430-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079317 Free PMC article.
-
Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins.J Virol. 2002 May;76(9):4267-74. doi: 10.1128/jvi.76.9.4267-4274.2002. J Virol. 2002. PMID: 11932392 Free PMC article.
-
Identification of a retroviral receptor used by an envelope protein derived by peptide library screening.Proc Natl Acad Sci U S A. 2007 Jun 26;104(26):11032-7. doi: 10.1073/pnas.0704182104. Epub 2007 Jun 20. Proc Natl Acad Sci U S A. 2007. PMID: 17581869 Free PMC article.
References
-
- Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Associates and Wiley-Interscience; 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

