HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2)
- PMID: 11533155
- PMCID: PMC114460
- DOI: 10.1128/JVI.75.19.8917-8926.2001
HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2)
Abstract
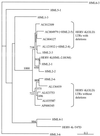
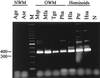
Sequences homologous to the human endogenous retrovirus (HERV) family HERV-K(HML-2) are present in all Old World primate species. A previous study showed that a central region of the HERV-K(HML-2) gag genes in Hominoidea species displays a 96-bp deletion compared to the gag genes in lower Old World primates. The more ancient HERV-K(HML-2) sequences present in lower Old World primates were apparently not conserved during hominoid evolution, as opposed to the deletion variants. To further clarify the evolutionary origin of the HERV-K(HML-2) family, we screened GenBank with the 96-bp gag-sequence characteristic of lower Old World primates and identified, to date, 10 human sequence entries harboring either full-length or partially deleted proviral structures, probably representing remnants of a more ancient HERV-K(HML-2) variant. The high degree of mutations demonstrates the long-time presence of these HERV-K(OLD) proviruses in the genome. Nevertheless, they still belong to the HML-2 family as deduced from dot matrix and phylogenetic analyses. We estimate, based on the family ages of integrated Alu elements and on long terminal repeat (LTR) divergence data, that the average age of HERV-K(OLD) proviruses is ca. 28 million years, supporting an integration time before the evolutionary split of Hominoidea from lower Old World primates. Analysis of HERV-K(OLD) LTR sequences led to the distinction of two subgroups, both of which cluster with LTRs belonging to an evolutionarily older cluster. Taken together, our data give further insight into the evolutionary history of the HERV-K(HML-2) family during primate evolution.
Figures




References
-
- Andersson M, Lindeskog M, Medstrand P, Westley B, May F, Blomberg J. Diversity of human endogenous retrovirus classII-like sequences. J Gen Virol. 1999;80:255–260. - PubMed
-
- Dangel A W, Baker B J, Mendoza A R, Yu C Y. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics. 1995;42:41–52. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

