RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome
- PMID: 11533204
- PMCID: PMC114509
- DOI: 10.1128/JVI.75.19.9415-9426.2001
RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome
Abstract
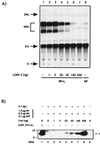
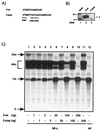
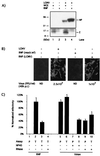
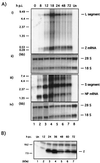
Arenaviruses have a bisegmented negative-strand RNA genome whose proteomic capability is limited to only four polypeptides, namely, nucleoprotein (NP), surface glycoprotein (GP) that is proteolytically processed into GP1+GP2, polymerase (L), and a small (11-kDa) RING finger protein (Z). The role of Z during the Lymphocytic choriomeningitis virus (LCMV) life cycle is poorly understood. We investigated the function of Z in virus transcription and replication by using a reverse genetic system for the prototypic arenavirus LCMV. This system involves an LCMV minigenome and the minimal viral trans-acting factors (NP and L), expressed from separated cotransfected plasmids. Cotransfection of the Z cDNA strongly inhibited LCMV minigenome expression. The effect required synthesis of Z protein; its magnitude was dose dependent and occurred with levels of Z protein substantially lower than those observed in LCMV-infected cells. Coexpression of Z did not prevent the encapsidation of plasmid supplied minigenome, but it affected both transcription and RNA replication similarly. Mutations in Z that unfolded its RING finger domain eliminated its inhibitory activity, but RING proteins not related to Z did not affect LCMV minigenome expression. Consistent with the minigenome results, cells transiently expressing Z exhibited decreased susceptibility to infection with LCMV.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

