Histone deacetylase activity represses gamma interferon-inducible HLA-DR gene expression following the establishment of a DNase I-hypersensitive chromatin conformation
- PMID: 11533238
- PMCID: PMC99796
- DOI: 10.1128/MCB.21.19.6495-6506.2001
Histone deacetylase activity represses gamma interferon-inducible HLA-DR gene expression following the establishment of a DNase I-hypersensitive chromatin conformation
Abstract
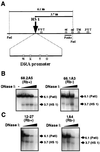
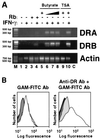
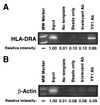
Expression of the retinoblastoma tumor suppressor protein (Rb) is required for gamma interferon (IFN-gamma)-inducible major histocompatibility complex class II gene expression and transcriptionally productive HLA-DRA promoter occupancy in several human tumor cell lines. Treatment of these Rb-defective tumor cell lines with histone deacetylase (HDAC) inhibitors rescued IFN-gamma-inducible HLA-DRA and -DRB mRNA and cell surface protein expression, demonstrating repression of these genes by endogenous cellular HDAC activity. Additionally, Rb-defective, transcriptionally incompetent tumor cells retained the HLA-DRA promoter DNase I-hypersensitive site. Thus, HDAC-mediated repression of the HLA-DRA promoter occurs following the establishment of an apparent nucleosome-free promoter region and before transcriptionally productive occupancy of the promoter by the required transactivators. Repression of HLA-DRA promoter activation by HDAC activity likely involves a YY1 binding element located in the first exon of the HLA-DRA gene. Chromatin immunoprecipitation experiments localized YY1 to the HLA-DRA gene in Rb-defective tumor cells. Additionally, mutation of the YY1 binding site prevented repression of the promoter by HDAC1 and partially prevented activation of the promoter by trichostatin A. Mutation of the octamer element also significantly reduced the ability of HDAC1 to confer repression of inducible HLA-DRA promoter activation. Treatment of Rb-defective tumor cells with HDAC inhibitors greatly reduced the DNA binding activity of Oct-1, a repressor of inducible HLA-DRA promoter activation. These findings represent the first evidence that HDAC activity can repress IFN-gamma-inducible HLA class II gene expression and also demonstrate that HDAC activity can contribute to promoter repression following the establishment of a DNase I-hypersensitive chromatin conformation.
Figures

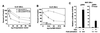
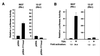
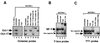
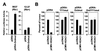

References
-
- Baskar S, Azarenko V, Garcia Marshall E, Hughes E, Ostrand-Rosenberg S. MHC class II-transfected tumor cells induce long-term tumor-specific immunity in autologous mice. Cell Immunol. 1994;155:123–133. - PubMed
-
- Baskar S, Clements V, Glimcher L, Nabavi N, Ostrand-Rosenberg S. Rejection of MHC class II-transfected tumor cells requires induction of tumor-encoded B7-1 and/or B7-2 costimulatory molecules. J Immunol. 1996;156:3821–3827. - PubMed
-
- Bennett M K, Tawny T N, Jyoti N A, Rosenfeld J M, Osborne T F. Co-stimulation of promoter for low-density lipoprotein receptor gene by sterol regulatory element-binding protein and Sp1 is specifically disrupted by the Yin-yang 1 protein. J Biol Chem. 1999;274:13025–13032. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
