Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4
- PMID: 11533240
- PMCID: PMC99798
- DOI: 10.1128/MCB.21.19.6515-6528.2001
Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4
Abstract
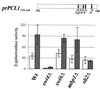
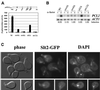
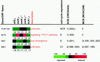
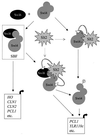
In Saccharomyces cerevisiae, the heterodimeric transcription factor SBF (for SCB binding factor) is composed of Swi4 and Swi6 and activates gene expression at the G(1)/S-phase transition of the mitotic cell cycle. Cell cycle commitment is associated not only with major alterations in gene expression but also with highly polarized cell growth; the mitogen-activated protein kinase (MAPK) Slt2 is required to maintain cell wall integrity during periods of polarized growth and cell wall stress. We describe experiments aimed at defining the regulatory pathway involving the cell cycle transcription factor SBF and Slt2-MAPK. Gene expression assays and chromatin immunoprecipitation experiments revealed Slt2-dependent recruitment of SBF to the promoters of the G(1) cyclins PCL1 and PCL2 after activation of the Slt2-MAPK pathway. We performed DNA microarray analysis and identified other genes whose expression was reduced in both SLT2 and SWI4 deletion strains. Genes that are sensitive to both Slt2 and Swi4 appear to be uniquely regulated and reveal a role for Swi4, the DNA-binding component of SBF, which is independent of the regulatory subunit Swi6. Some of the Swi4- and Slt2-dependent genes do not require Swi6 for either their expression or for Swi4 localization to their promoters. Consistent with these results, we found a direct interaction between Swi4 and Slt2. Our results establish a new Slt2-dependent mode of Swi4 regulation and suggest roles for Swi4 beyond its prominent role in controlling cell cycle transcription.
Figures




References
-
- Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. Boston, Mass: John Wiley & Sons, Inc.; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
