Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells
- PMID: 11533249
- PMCID: PMC99807
- DOI: 10.1128/MCB.21.19.6615-6625.2001
Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells
Abstract
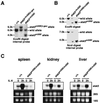
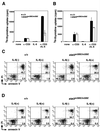
Signal transducer and activator of transcription 3 (STAT3) mediates signals of various growth factors and cytokines, including interleukin-6 (IL-6). In certain IL-6-responsive cell lines, the stat3 gene is autoregulated by STAT3 through a composite IL-6 response element in its promoter that contains a STAT3-binding element (SBE) and a cyclic AMP-responsive element. To reveal the nature and roles of the stat3 autoregulation in vivo, we generated mice that harbor a mutation in the SBE (stat3(mSBE)). The intact SBE was crucial for IL-6-induced stat3 gene activation in the spleen, especially in the red pulp region, the kidney, and both mature and immature T lymphocytes. The SBE was not required, however, for IL-6-induced stat3 gene activation in hepatocytes. T lymphocytes from the stat3(mSBE/mSBE) mice were more susceptible to apoptosis despite the presence of IL-6 than those from wild-type mice. Consistent with this, IL-6-dependent activation of the Pim-1 and junB genes, direct target genes for STAT3, was attenuated in T lymphocytes of the stat3(mSBE/mSBE) mice. Thus, the tissue-specific autoregulation of the stat3 gene operates in vivo and plays a role in IL-6-induced antiapoptotic signaling in T cells.
Figures




Similar articles
-
Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice.J Immunol. 1998 Nov 1;161(9):4652-60. J Immunol. 1998. PMID: 9794394
-
Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein.J Biol Chem. 1998 Mar 13;273(11):6132-8. doi: 10.1074/jbc.273.11.6132. J Biol Chem. 1998. PMID: 9497331
-
Interleukin-6 induces transcriptional activation of vascular endothelial growth factor (VEGF) in astrocytes in vivo and regulates VEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1.Int J Cancer. 2005 Jun 10;115(2):202-13. doi: 10.1002/ijc.20871. Int J Cancer. 2005. PMID: 15688401
-
Multi-functional roles of Stat3 revealed by conditional gene targeting.Arch Immunol Ther Exp (Warsz). 2001;49(4):279-83. Arch Immunol Ther Exp (Warsz). 2001. PMID: 11726030 Review.
-
IL-6-regulated transcription factors.Int J Biochem Cell Biol. 1997 Dec;29(12):1401-18. doi: 10.1016/s1357-2725(97)00063-0. Int J Biochem Cell Biol. 1997. PMID: 9570135 Review.
Cited by
-
T helper cells from aggressive periodontitis patients produce higher levels of interleukin-1 beta and interleukin-6 in interaction with Porphyromonas gingivalis.Clin Oral Investig. 2014 Sep;18(7):1835-43. doi: 10.1007/s00784-013-1162-5. Epub 2013 Dec 19. Clin Oral Investig. 2014. PMID: 24352581
-
NFκB and STAT3 synergistically activate the expression of FAT10, a gene counteracting the tumor suppressor p53.Mol Oncol. 2014 May;8(3):642-55. doi: 10.1016/j.molonc.2014.01.007. Epub 2014 Jan 24. Mol Oncol. 2014. PMID: 24518302 Free PMC article.
-
Quinazoline Ligands Induce Cancer Cell Death through Selective STAT3 Inhibition and G-Quadruplex Stabilization.J Am Chem Soc. 2020 Feb 12;142(6):2876-2888. doi: 10.1021/jacs.9b11232. Epub 2020 Jan 28. J Am Chem Soc. 2020. PMID: 31990532 Free PMC article.
-
Inherited human ZNF341 deficiency.Curr Opin Immunol. 2023 Jun;82:102326. doi: 10.1016/j.coi.2023.102326. Epub 2023 Apr 18. Curr Opin Immunol. 2023. PMID: 37080116 Free PMC article. Review.
-
Who regulates whom: ZNF341 is an additional player in the STAT3/TH17 song.Sci Immunol. 2018 Jun 15;3(24):eaat9779. doi: 10.1126/sciimmunol.aat9779. Sci Immunol. 2018. PMID: 29907692 Free PMC article.
References
-
- Akira S, Nishio Y, Inoue M, Wang X J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. - PubMed
-
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. - PubMed
-
- Catlett-Falcone R, Landowski T H, Oshiro M M, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna J L, Nunez G, Dalton W S, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
